Figure 1.
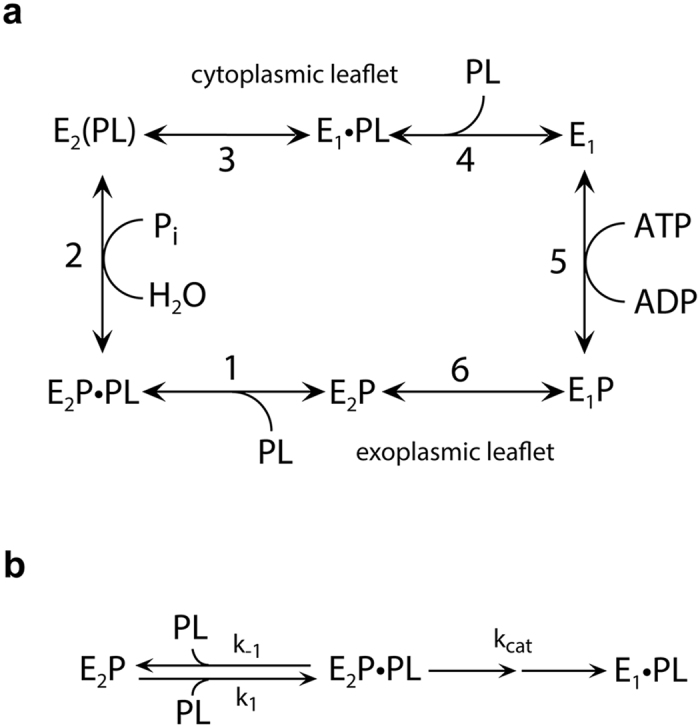
Flippase reaction cycle. (a) The scheme is based on functional similarities to the ion transporting P-type ATPases15. E1, E1P, E2P, and E2 are the major enzyme conformational states, the “P” indicating covalently bound phosphate. Reactions are numbered 1–6. The phospholipid substrate, PL, enters the cycle from the exoplasmic leaflet of the lipid bilayer by binding to the E2P phosphoenzyme (Reaction 1), thereby stimulating the dephosphorylation (Reaction 2) and release of the lipid toward the cytoplasmic leaflet (Reaction 4) as a consequence of the E2 to E1 transition (Reaction 3). The transition to E1 allows the enzyme to become rephosphorylated (Reaction 5). The stimulation of dephosphorylation by the lipid substrate is analogous to the activation of Na+,K+-ATPase dephosphorylation by K+ binding to E2P from the extracellular side of the membrane2. In further analogy with the modes of binding and transport of K+ by the Na+,K+-ATPase, the parentheses in E2(PL) indicate that the PL is bound to the protein in an occluded state, where the protein conformational change to E1 is required before the PL can be released toward the cytoplasmic side. In E2P·PL and E1·PL the · indicates that PL in these complexes is able to dissociate (to the exoplasmic leaflet and the cytoplasmic leaflet, respectively). (b) Rate constants relating to the rate-limiting dephosphorylation reaction and subsequent conformational change. Under the experimental conditions applied here, the dephosphorylation is essentially irreversible, because the concentration of inorganic phosphate in the medium is low.
