Fig. 4.
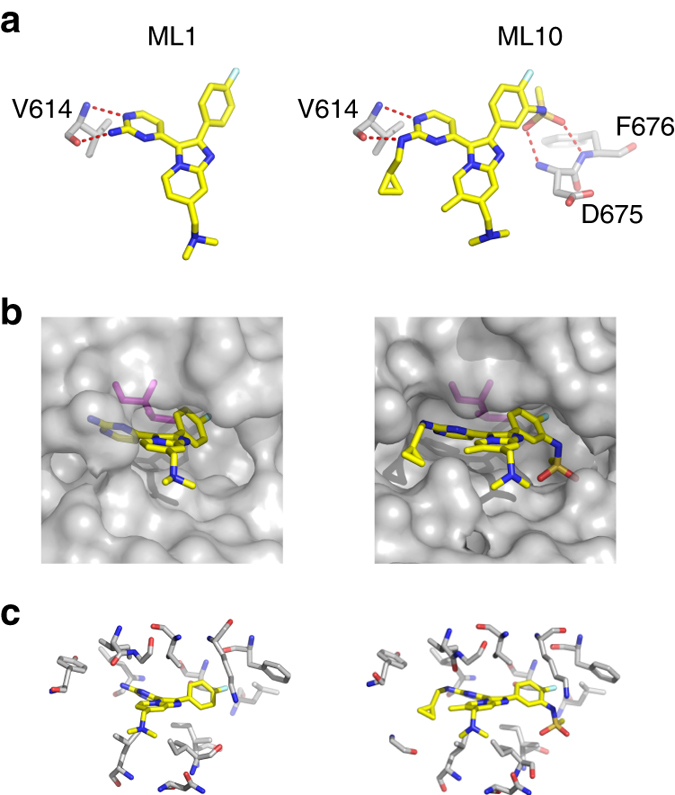
Side-by-side comparison with key features in the PvPKG/ML1 and PvPKG/ML10 co-structures. a Hydrogen bonds provide key interactions: At the hinge of the kinase domain, V614 forms hydrogen bonds with the amino-pyrimidine group of both ML1 (left) and ML10 (right). In addition, ML10 extends a sulfonamide moiety to form hydrogen bonds with D675 and F676, resulting in significantly greater inhibitory potency than ML1. b Gatekeeper confers specificity: Both ML1 (left) and ML10 (right) extend a fluorophenyl group to occupy a hydrophobic pocket adjacent to T611 (magenta). This explains in part the specificity of the inhibitors as a residue with a longer side-chain may potentially clash with this functional group. c Hydrophobic network enhances inhibitory potency: Plasmodium PKG also interacts with both ML1 (left) and ML10 using a network of hydrophobic residues (see Supplementary Fig. 13 for the same figure with amino-acid labels)
