Figure 1.
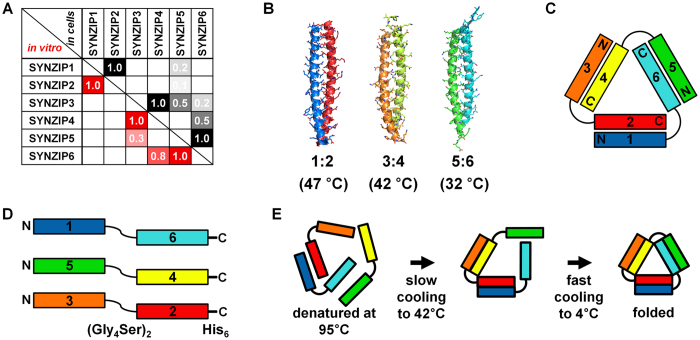
Orthogonally interacting SYNZIPs underlie the design of a protein nanotriangle. (A) Pairwise interaction profiles of SYNZIPs (1 to 6) determined by protein microarrays (in vitro, lower left)20 and the yeast two-hybrid assays (in cells, upper right)19. Assay scores were normalized to a scale where 1.0 indicates tight binding. The corresponding K D values are <10, <30, and <15 nM for 1:2, 3:4, and 5:6, respectively, and are approximated to be >400 nM for 3:5, and 4:6 19. (B) Crystal structures of 1:2 (PDB ID: 3HE5) and 5:6 (PDB ID: 3HE4)20, and a model of 3:4; melting temperatures are given in parentheses. (C) Topology of the designed heterotrimeric protein nanotriangle composed of the orthogonal SYNZIPs. (D) The composition of individual linked-SYNZIP fusion proteins that self-assemble into the nanotriangle shown in panel C. (E) A two-step thermal annealing process for assembly of the linked-SYNZIP fusion proteins, and illustration of desired assemblies at each step.