Fig. 4.
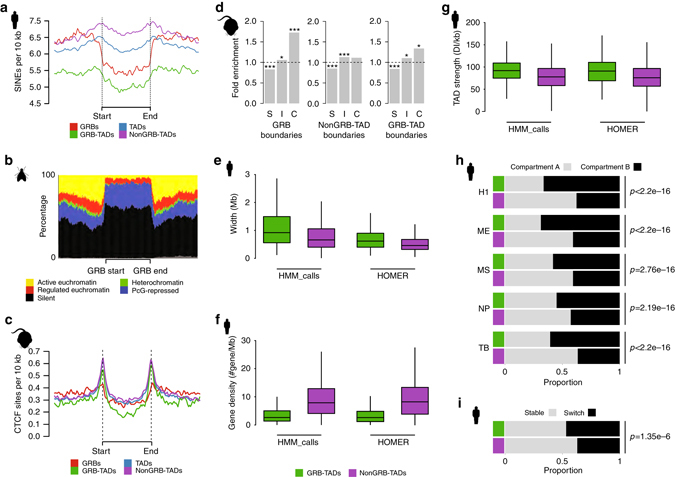
Several sets of features distinguish between TADs associated with extreme conservation from those without. a Depletion of SINE elements within GRB-TADs compared to non-GRB-TADs (using H1 HOMER TADs) reflects the selective constraint on these regions against the retention of repeat element insertion. b GRBs in Drosophila Kc167 cells are mainly associated with inactive (black) and Polycomb-repressed chromatin (blue) and represent functionally coherent regions. Active and regulated chromatin correspond to different types of euchromatin. There appears to be a change in the proportion of constitutively active chromatin (yellow) at the boundary regions identified as GRBs. c CTCF sites are depleted within GRBs. CTCF sites per 10 kb plotted across GRBs and flanking regions of equivalent size to the GRBs normalised to show signal relative to GRB boundaries and Loess-smoothed. d Enrichment for different patterns of CTCF binding at different genomic features (Specific, Intermediate, Constitutive). Constitutive CTCF peaks are enriched within 10 kb of both GRBs and GRB-TAD boundaries (binomial test p-values: *p < 0.05, **p < 0.01, ***p < 0.001). e Distribution of TAD width reveals GRB-TADs are significantly longer than non-GRB-TADs identified in human H1 cells using either HOMER (median width 620 kb vs. 460 kbp, p < 1e−6) or HMM_calls (median width 920 vs. 680 kb, p < 1e−6). f Human H1 GRB-TADs are associated with lower protein-coding gene density than non-GRB-TADs identified using HOMER (median no. genes 2.63 vs. 8.33 p < 1e–6) or HMM_calls (median no. of genes 2.65 vs. 8.33 p < 1e−6). g GRB-TADs are significantly stronger than non-GRB-TADs identified in human H1 cells using HOMER (median strength 90.88 vs. 75.81, p < 1e−6) or HMM_calls (median strength 91.77 vs. 77.68, p < 1e−6). h GRB-TADs (HOMER) are preferentially associated with compartment B in all of the lineages investigated. i GRB-TADs are more likely to switch compartment in at least one of the five lineages investigated (i.e., A–B or B–A) than non-GRB-TADs
