Fig. 5.
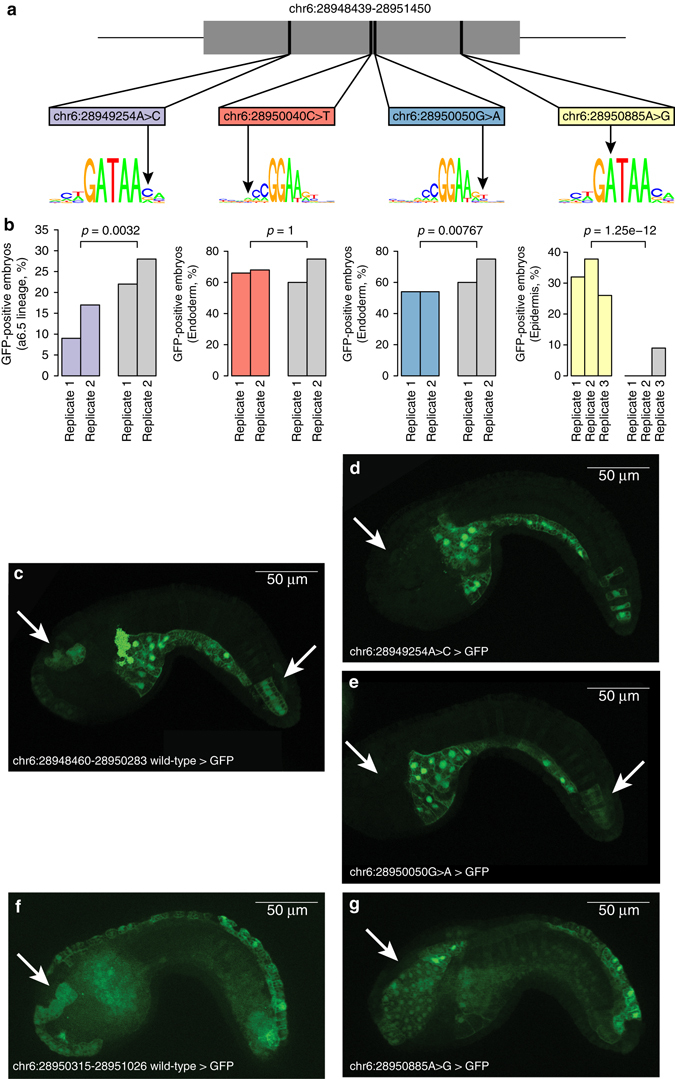
In vivo validation of putative driver mutations in DHS chr6:28948439-28951450. a Shown are the locations of the four mutations in GATA and ETS transcription factor binding sites that were tested for function. b Barplots showing downregulation in anterior neural plate (a6.5 lineage, chr6:28949254 A > C), no change in endoderm (chr6:28950040 C > T), downregulation in endoderm (chr6:28950050 G > A) and overexpression in head epidermis (chr6:28950885 A > G). p-values were calculated using Fisher’s exact test. (c–g) Images showing tailbud stage C. intestinalis embryos (8 h post fertilization at 21 degrees Celsius) electroporated with (c, f) indicated reference enhancer > GFP or (d, e, g) indicated mutated enhancer > GFP. d Decreased expression in the anterior neural plate is observed when the enhancer is mutated (chr6:28949254 A > C); e Enhancer activity is decreased in anterior neural plate and secondary notochord in presence of mutation chr6:28950050 G > A. c is the wild-type enhancer for both d and e. g Increased expression in the epidermis is observed when the enhancer is mutated (chr6:28950885A>G)
