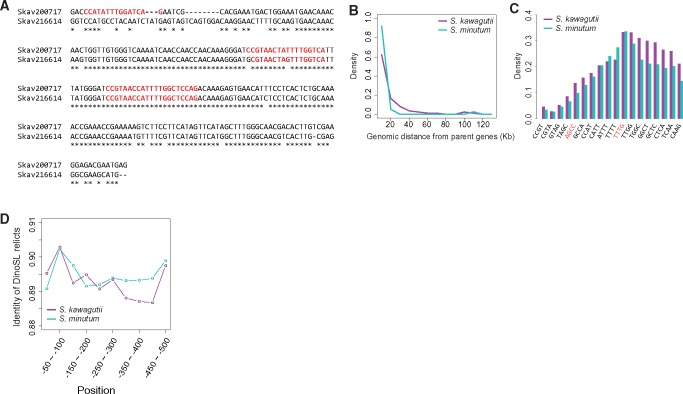
Fig. 1.
—Retroposition in Symbiodinium genome sequences retaining DinoSL (CCGTAGCCATTTTGGCTCAAG) motifs. (A) Alignment of a retrogene (Skav200717) and its parent (Skav216614), both of which have undergone repeated retroposition events. Red colored sequences show DinoSL relicts. (B) Distance between retrogenes and their parents in S. kawagutii (purple line) and S. minutum (cyan line). (C) DinoSL motif “TTTT,” “TTTG,” and “TTGG” are preferentially retained in all the detected DinoSL relicts in both S. kawagutii (purple bars) and S. minutum (cyan bars). (D) Identities of DinoSL relicts of S. kawagutii (purple line) and S. minutum (cyan line) as a function of upstream distance from the start codon. Higher identities of DinoSLs are seen between −50 and −100 bp where the promoter is expected to be located.