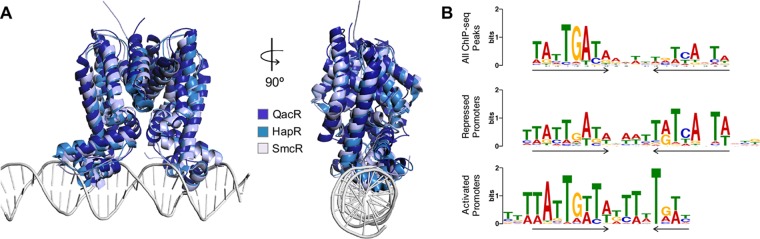
FIG 2.
LuxR/HapR homolog structural and DNA binding properties. (A) Superimposed crystal structures of the TetR proteins QacR, HapR, and SmcR (32, 33, 84). Structures were superimposed with DaliLite (142) and the figures created with PyMOL. The DNA-bound QacR dimer superimposed with HapR or SmcR both resulted in a root mean square deviation (RMSD) of 3.1. Superimposition of HapR to SmcR results in an RMSD of 1.5. HapR is shown in cyan (PDB: 2PBX), SmcR is shown in light blue (PDB: 3KZ9), and for the QacR-DNA structure, QacR is shown in dark blue and the DNA in gray (PDB: 1JT0) (32, 33, 84). (B) LuxR DNA binding motifs from ChIP-seq data grouped by the location of the binding site peak. Top, all LuxR DNA binding peaks from ChIP-seq; middle, peaks in promoters of repressed genes; bottom, peaks in promoters of activated genes. The arrows indicate dyad symmetry in the binding site. Image reproduced with permission (74).