Figure 4.
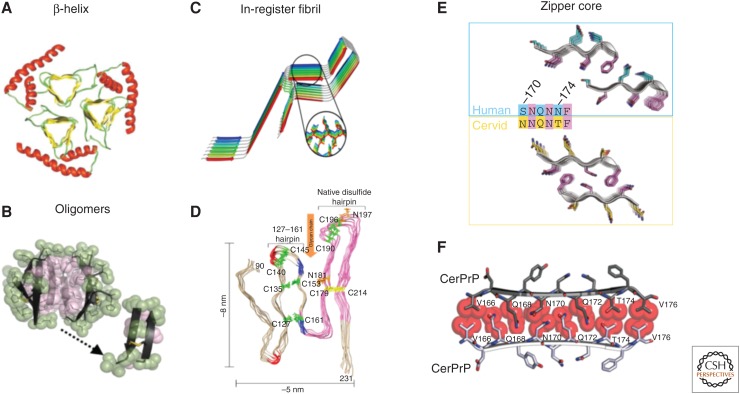
Proposed models for PrPSc. (A) A triangular β-helix model of PrPSc was based on a transmission electron microscopy (TEM) image. (Courtesy of Govaerts et al. 2004; reprinted, with permission, from the National Academy of Sciences, U.S.A. © 2001.) (B) An atomic structure for a PrP oligomer was determined from pairs of naturally disulfide-linked segments with PrP sequences. (Courtesy of Apostol et al. 2013; reprinted, with permission, from the American Chemical Society © 2013.) (C,D) In-register fibril models have been obtained from different experiments. (C) An in-register stacking model was proposed based on electron paramagnetic resonance (EPR) of human PrP fibrils converted in vitro. (Courtesy of Cobb et al. 2007; reprinted, with permission, from the National Academy of Sciences, U.S.A. © 2007.) (D) Another parallel in-register β-sheet architecture using solid-state NMR spectroscopy of recombinant PrP amyloid fibrils that were seeded with PrPSc. (Courtesy of Groveman et al. 2014; adapted, with permission, © 2014, by the American Society for Biochemistry and Molecular Biology.) (E,F) Zipper structures of segments in or near the β2–α2 loop highlight its importance in conversion and transmission. (E) Microcrystal structures of human (cyan, top) and cervid (yellow, lower) β2–α2 loop segments belong to different classes of steric zippers, supporting the idea that side-chain mismatches may account for species barriers in prion disease. (Courtesy of Apostol et al. 2011; adapted, with permission, © 2011 American Chemical Society.) (F) Zipper model of the β2–α2 loop highlights tight, complementary side-chain interactions that are required for efficient prion conversion. (Courtesy of Kurt et al. 2014b; adapted, with permission, © 2014, by the American Society for Biochemistry and Molecular Biology.)