Fig. 1.
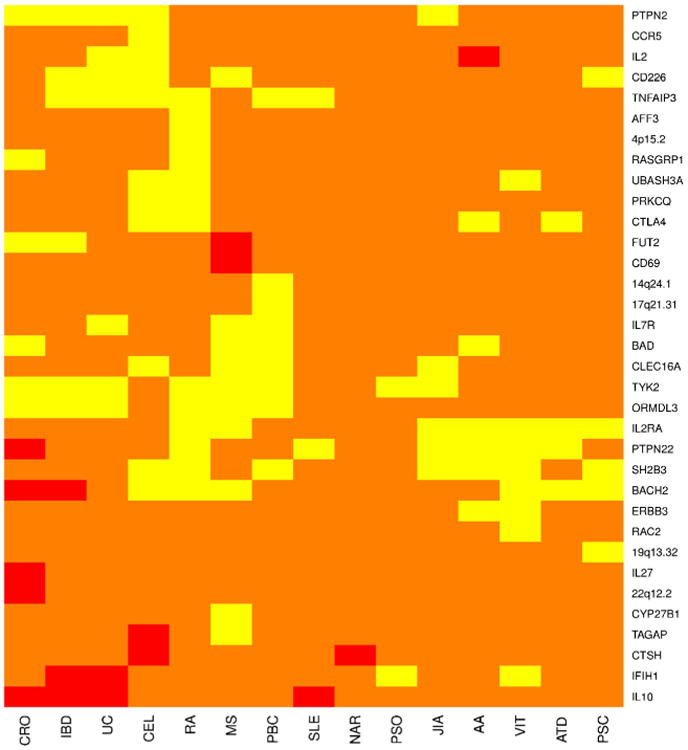
A heatmap showing the shared genetics of 15 autoimmune diseases with T1D at 34 non-MHC loci gathered from information contained at the Immunobase website (www.immunobase.org). All relevant publications can be found in a curated list at the Immunobase website. The regions are named for potential causative genes located at the locus and do not represent the actual causal gene. The regions are defined by the location of the index SNP at the locus and extending out +/–0.1 cM from that location. Yellow indicates that a SNP in this region has odds ratios that are in the same direction for both T1D and the disease in question or the direction of effect was unknown for one the diseases. Red indicates that a SNP in this region has odds ratios that are opposing each other in direction for both diseases. Orange represents a region with no known SNP sharing significance with T1D at this time. For a SNP to be considered shared it must be genome-wide significant (p<5.0×10–8) in one of the diseases and at least suggestive of significance (p<1.0×10– 5) in the other diseases. AA=alopecia areata, ATD=autoimmune thyroid disease, CEL=celiac disease, CRO=Crohn's disease, JIA=juvenile idio-pathic arthritis, MS=multiple sclerosis, PBC=primary biliary cirrhosis, PSO=psoriasis, RA=rheumatoid arthritis, SLE=systemic lupus erythematosis, T1D=type 1 diabetes, UC=ulcerative colitis, IBD=in-flammatory bowel disease, NAR=narcolepsy, PSC=primary sclerosing cholangitis, VIT=vitiligo
