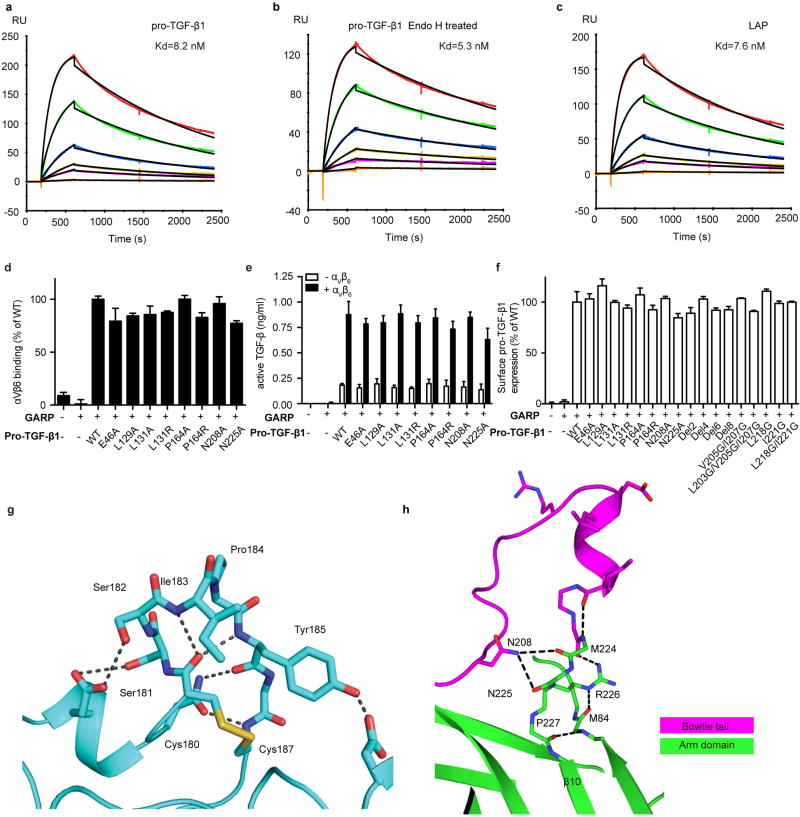
Figure S4. Binding, activation, and structural details.
(a–c) Surface plasmon resonance measurements of integrin αVβ6 headpiece binding to surface immobilized furin-mutant pro-TGF-β1 (a), Endo H treated furin-mutant pro-TGF-β1 (b), and the untreated prodomain expressed in absence of the GF (latency-associated peptide, LAP) (c). Curves are colored red, 100 nM; dark green, 50 nM; blue, 20 nM; yellow, 10 nM; magenta, 5 nM; orange, 1 nM. The global fit to the 1:1 binding model is in black. Kd and chi-square values from fits are shown. (d) Binding in presence of Mg2+ of FITC-αVβ6 to WT or mutant pro-TGF-β1/GARP HEK293T co-transfectants as specific mean fluorescence intensity (MFI) (% of WT). The mutated residues lie on the arm domain outside the RGDLATI motif. (e) Activation of TGF-β1 by HEK293T cells co-tranfected with pro-TGF-β1 and GARP, with or without αV and β6, assayed with luciferase reporter cells, and standardized with purified TGF-β1. The mutated residues lie on the arm domain outside the RGDLATI motif. (f) Expression of WT and mutant pro-TGF-β1/GARP complexes on 293T transfectants determined by immunofluorescence flow cytometry with TW7-28G11 antibody. Values represent the mean of 3 individual tests and the error bars represent the s. d.. (g) Hydrogen bonds within SDL2 in the β6 βI domain. (h) The hydrogen bond network in the 4 residues, M224-P227, that link the bowtie tail to the β10 strand in the integrin-bound pro-TGF-β1 arm domain.
Binding of integrin αVβ6 to pro-TGF-β1 does not liberate the GF4. The experiments in Extended Data Fig. S4 panels a–c address the question of whether integrin binding loosens the prodomain’s grip on the GF. Destabilization of the binding energy for the GF would require an identical stabilization of integrin binding to the isolated prodomain compared to the pro-complex; however, this is ruled out by the equivalent dissociation constants for integrin αVβ6 binding to the prodomain (7.6 nM, panel c) and pro-complex (8.2 nM, panel a). Thus, in the absence of force, there is no propagation of conformational change from the integrin binding site to the prodomain:GF interface that lowers affinity for ligand.
The β6 SDL2 backbone is supported by many backbone hydrogen bonds (panel g), correlating with the finding that in the open β6 conformation determined here, SDL2 is essentially identical in backbone to the previous closed conformation bound to the TGF-β3 peptide 5. In contrast, SDL2 in β2 moves, even in intermediate headpiece opening 22.