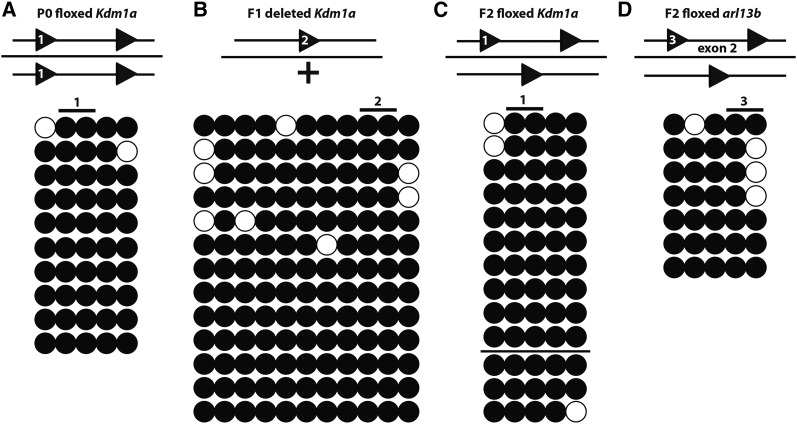
Figure 5.
Bisulfite DNA methylation analysis of the LoxP recognition site. In vivo bisulfite analysis of the two CpG dinucleotides (Figure 1A) in the LoxP recognition sequence (diagrammed above), as well as CpG residues from the flanking region, in the Kdm1a P0 floxed allele (marked with 1) prior to recombination (A) (N = 5 mice), the Kdm1a F1 deleted allele (marked with 2) after recombination (B) (N = 5 mice), Kdm1a F2 floxed alleles (marked with 1) that do not recombine (nine lollipop rows above the horizontal black line), and Kdm1a F2 floxed alleles recombine normally (four lollipop rows below the horizontal black line) (C) (N = 13 total mice), and the Arl13b P0 floxed allele (marked with 3) (D) (N = 3 mice). Open circles indicate unmethylated CpG residues. Closed circles indicate methylated CpG residues.