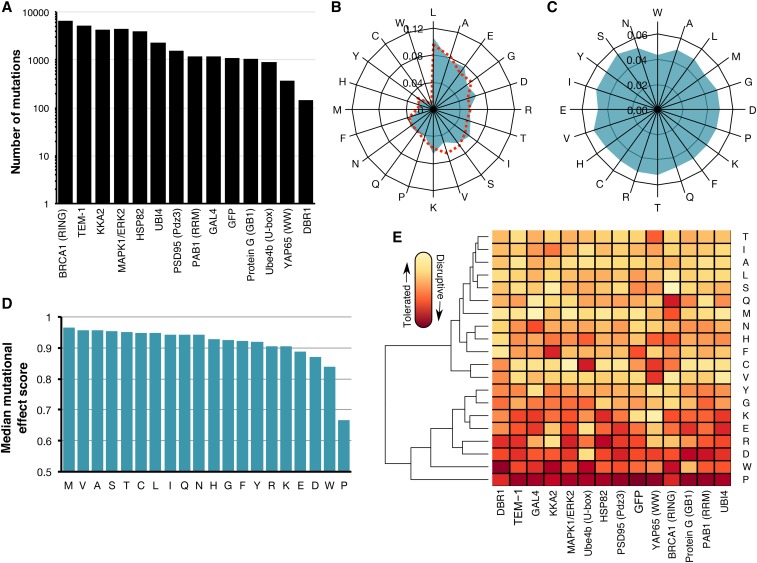
Figure 1.
Large-scale mutagenesis data from 14 proteins. (A) The number of single amino acid mutations with effect scores in each of the 14 proteins is shown. (B) A radar plot shows the relative frequency of occurrence of each amino acid in the wild-type sequences of the 14 proteins (blue) or in 554,515 proteins in the UniProt Knowledgebase (Magrane and UniProt Consortium 2011) (dashed red). (C) A radar plot shows the relative frequency of each of the 20 amino acid substitutions in the large-scale mutagenesis data sets for all 14 proteins. (D) The median mutational effect score of each amino acid substitution is shown for 34,373 mutations at 2236 positions in all 14 proteins. (E) A heat map shows the median mutational effect score of each amino acid substitution for each protein separately. Yellow indicates tolerated substitutions while orange indicates disruptive substitutions. Amino acids and proteins were ordered according to similarity using hierarchical clustering with the hclust function from the heatmap2 package in R.