Figure 4.
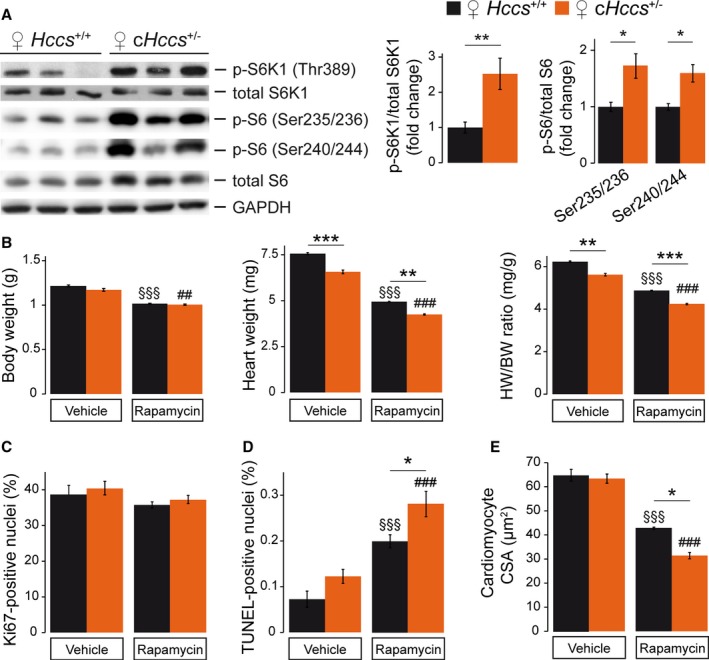
Impact of fetal mTORC1 inhibition on a mouse model of prenatal compensatory cardiac growth. A, Western blots illustrating the phosphorylation status of S6K1 and S6 ribosomal protein revealed enhanced mTORC1 activity in neonatal (P1) cHccs +/− hearts compared to littermate controls (densitometric quantification for S6K1: Hccs +/+ n=15, cHccs +/− n=14, for S6: n=6 per group). B, Body weight (BW) of rapamycin‐treated neonates was significantly reduced compared to vehicle‐treated animals, but no difference was observed between genotypes. Rapamycin‐treated Hccs +/+ and cHccs +/− neonates demonstrated similar reductions in heart weight (HW) and HW/BW ratio compared to vehicle‐treated animals. Note that the reduced HW and HW/BW ratio in cHccs +/− compared to Hccs +/+ newborns reported previously12 persisted after prenatal mTORC1 inhibition (vehicle groups n=9, rapamycin groups n=10). C, Quantification of Ki67‐positive nuclei revealed unchanged proliferation rates within the LV myocardium of vehicle‐ and rapamycin‐treated hearts and between genotypes (vehicle groups n=6, rapamycin Hccs +/+ n=12, rapamycin cHccs +/− n=13). D, Quantification of TUNEL‐positive nuclei revealed significantly increased apoptosis in hearts after prenatal mTORC1 inhibition, with apoptosis in rapamycin‐treated cHccs +/− hearts being significantly higher than in rapamycin‐treated Hccs +/+ controls (vehicle Hccs +/+ n=4, all other groups n=6). E, Prenatal rapamycin treatment significantly reduced cardiomyocyte cross‐sectional area (CSA) in both genotypes, and CSA in rapamycin‐treated cHccs +/− hearts was significantly smaller compared to rapamycin‐treated Hccs +/+ controls (vehicle groups n=5, rapamycin groups n=3). B through E, Note that data used for Hccs +/+ animals are the same as depicted in Figures 2 and 3, respectively. (*P<0.05, **P<0.01, ***P<0.001; §§§ P<0.001 vs vehicle Hccs +/+; ## P<0.01, ### P<0.001 vs vehicle cHccs +/−). mTORC1 indicates mechanistic target of rapamycin complex 1; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
