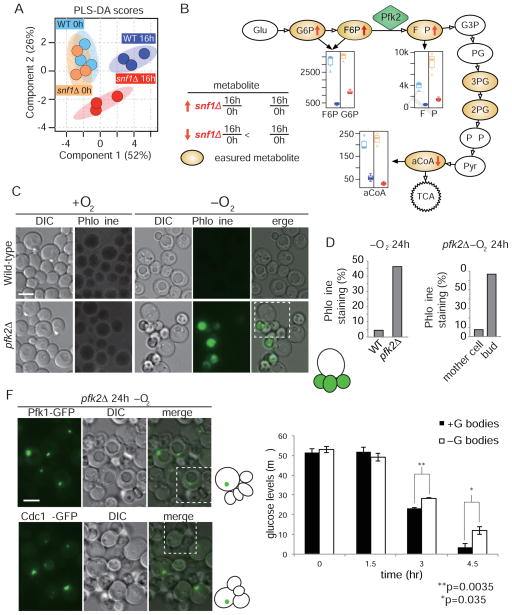
Figure 4. Snf1 regulates glycolytic activity under hypoxic conditions.
(A) Hypoxia and snf1 deletion result in remodeling of metabolic pathways. Cross-validated partial least squares discriminant analysis (PLS-DA) score plot (R2Y=0.936; Q2=0.454) shows separation of wild-type and snf1Δ samples at time 0 and after 16 h hypoxic treatment. Shaded ovals represent 95% confidence intervals. Solid circles represent individual replicates. (B) Four key metabolites in the glycolysis pathway contribute to the metabolic differences observed in (A). Boxplot colors represent samples as in (A). (C–E) Phloxine B staining indicates high bud cell death in pfk2Δ cells after 24 h hypoxia. Dead cells were defined as those with fluorescence signal in the FITC channel. Quantification in (D, E) is based on N>300 cells per strain. Cartoon is of mother cell and daughters in the dotted box, green indicates Phloxine B. (F) Pfk1p-GFP and Cdc19p-GFP form granules in pfk2Δ cells. C, F scale bar 5 μm. Cartoon is of mother cell and daughters in the dotted box, green indicates GFP-tagged protein. (G) Cells with G bodies have significantly less glucose than cells lacking G bodies by t-test at t = 3 h (p=0.0035) and t = 4.5 h (p=0.035). Error bars represent SEM for three biological replicates in each condition. [See also Figure S7]