Figure 2. Structural features of the FGF9mut-FGFR1cecto/252R complex.
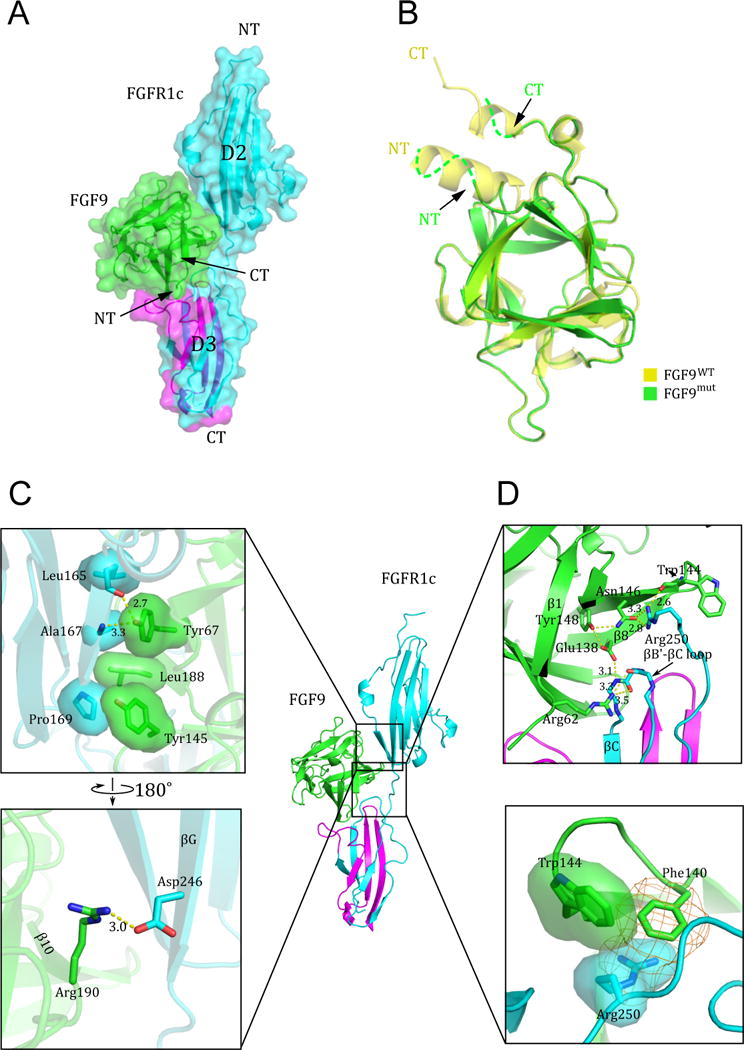
(A) The FGF9mut-FGFR1cecto/P252R complex structure in both cartoon and surface representations in traditional “front view” orientation. The receptor is colored cyan except for its alternatively spliced region in D3, which is colored magenta. The ligand (in green) engages D2, D3, and D2-D3 linker of the receptor. (B) Superimposition of of free (unbound) FGF9 (PDB ID: 1IHK) (Plotnikov et al., 2001) onto the receptor-bound FGF9 shows disordering of the FGF9 N-terminus upon receptor binding. NT and CT denote N- and C-termini of FGF9 and FGFR1c. Arrows point to the N- and C-terminus of receptor-bound FGF9 to emphasize their disordering in the receptor-bound state. (C) The interface between FGF9 and FGFR1c D2 domain. Hydrophobic interactions between Tyr-67, Tyr-145, and Leu188 of FGF9 and Leu-165, Ala-167, and Phe-169 in D2 of receptor (upper box) are highly conserved, whereas the hydrogen bonding contact between Arg-190 of FGF9 and Asp-246 of FGFR1c (lower box) is seen only in a subset of FGF-FGFR complexes. (D) The interface between FGF9 and FGFR1c D2-D3 linker and the constant (unspliced) region of D3. The upper box shows the conserved network of hydrogen bonds between Trp-144 and Asn-146 FGF9 and Arg-250 of the receptor D2-D3 linker, and between Glu-138 (in β8) and Arg-62 (in β1) of FGF9 with the backbone of the βB′-βC loop in the ‘unspliced’ constant region of the D3 domain. The π-cation (orange mesh) and hydrophobic interactions (transparent surface) between Phe-140 and Trp-144 of FGF9 and the side chain of Arg-250 are unique to the FGF9 subfamily (lower box). See also Figure S1.
