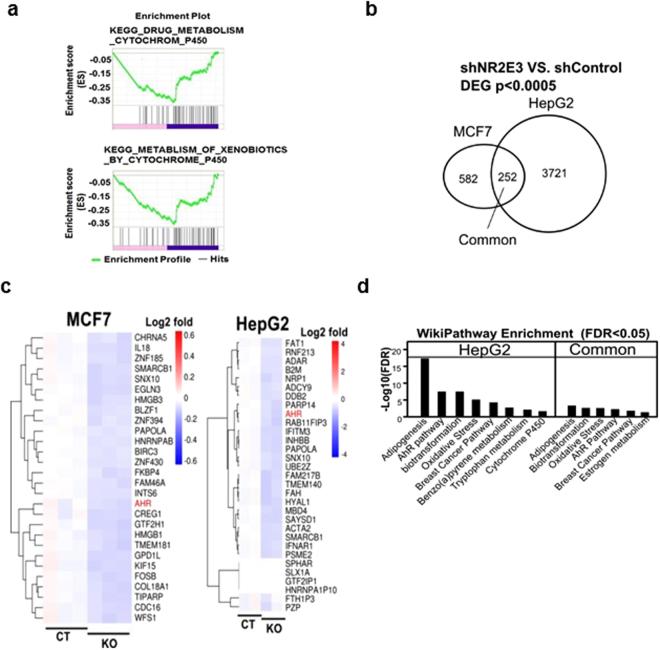
Figure 1.
Identification of AHR as a novel target of NR2E3 by RNA sequencing (RNA-seq) and bioinformatics analyses. (a) Gene set enrichment analysis by employing RNA-seq data derived from small hairpin RNA control (shCT) and NR2E3-depleted (shNR2E3) HepG2 cell RNA lysate showed that NR2E3 gene networks are associated with drug metabolism involving cytochrome p450 and metabolism of xenobiotics by cytochrome p450 signaling pathways. (b) Venn diagram revealed 252 differentially expressed genes (DEGs) commonly regulated by NR2E3 depletion in both MCF-7 and HepG2 cells (p < 0.0005). (c) Heat map of the 253 common genes indicates that AHR expression (red) is markedly downregulated in both MCF-7 and HepG2 cells. (d) WikiPathway analysis using the common gene set showed the association of NR2E3-regulated genes with AHR-related signaling pathways (FDR < 0.05).