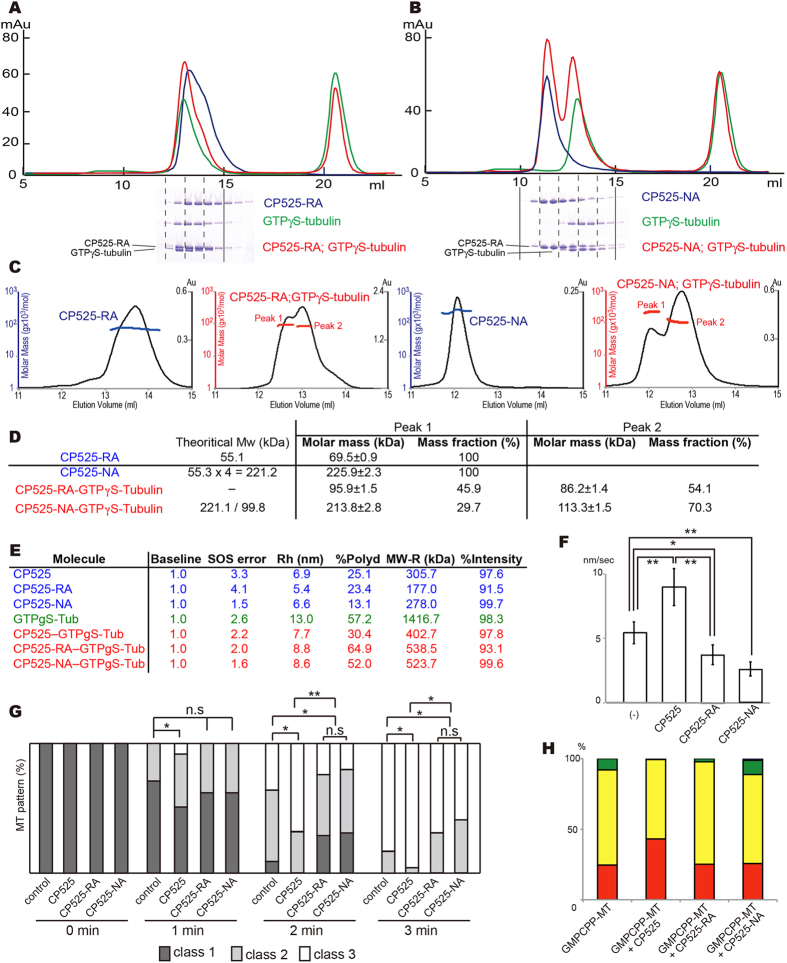
Figure 6.
Structure-guided CP525 interface mutants abolish CRMP2-tubulin binding. (A,B) Size exclusion chromatography of CRMP2 mutants (blue), GTP-γS-tubulin-dimer (green), and CRMP2-mutants with GTP-γS-tubulin-dimer (red), shown with SDS-PAGE results. (A) CP525-RA. (B) CP525-NA. (C) SEC-MALS analyses of CP525 mutants and the mixtures with GTP-γS-tubulin. (D) The molar mass determined by SEC-MALS experiments. Theoretical molecular weights calculated from the amino acid sequence were also shown for comparison. (E) DLS results of CP525, CP525 mutants, GTP-γS-tubulin-dimer, and CP525/CP525 mutants with GTP-γS-tubulin-dimer. (F) Microtubule growth rate without CRMP2 (n = 7), in the presence of CP525 (n = 11), CP525-RA (n = 9), and CP525-NA (n = 9). Error bars represent the S.D. *P < 0.01; **P < 0.001. t-test. (G) Time course of microtubule polymerization after the cold-induced depolymerization without CRMP2, with CP525 wild-type or with CP525 interface mutants. A representative result from three independent experiments is shown. n = 100 cells for each transfection. *P < 0.05; **P < 0.01, compared to control. Chi-square test with Bonferroni correction. Classification is explained in Fig. 2B. (H) Percentage of end types observed. GMPCPP-microtubule (n = 187); GMPCPP-microtubule+CP525 (n = 184); GMPCPP-microtubule+CP525-RA (n = 200); GMPCPP-microtubule+CP525-NA (n = 198).