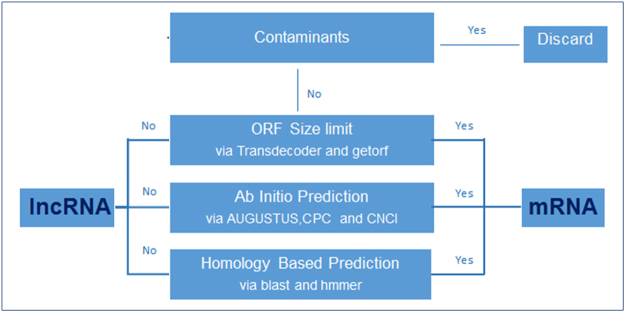
Figure 1.
The pipeline for the identification and annotation of both coding transcripts and lncRNAs. Before the identification of both coding transcripts and lncRNAs, the organellar transcripts and small RNAs were eliminated from the transcripts which were all together named as ‘contaminants’. The open reading frame (ORF) of transcripts were detected with two different softwares, Transdecoder and EMBOSS:getorf, since the presence of functional ORF is the most important criteria for identification of both coding transcripts and lncRNAs. Coding potential of transcripts was calculated with two different programs, CNCI and CPC, in addition to the online gene prediction tool, AUGUSTUS. Additionally, the homology between transcripts and known proteins sequences were inspected, together with the functional protein domains.