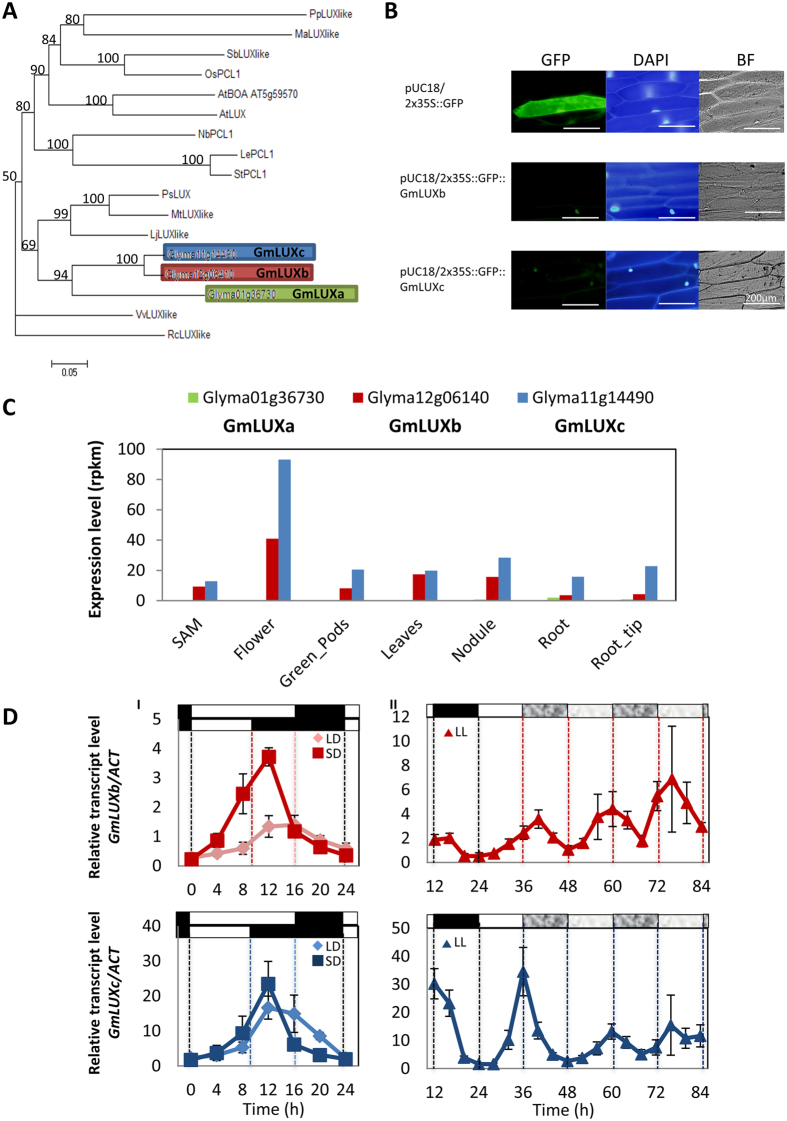
Figure 1.
Characterisation of soybean LUX ARRHYTHMO (LUX). (A) Phylogenetic analysis of soybean LUX proteins. Three soybean homologues were identified in the soybean genome and designated as GmLUXa, GmLUXb, and GmLUXc. The tree is drawn to given scale, and the bootstrap values are shown at each node. (B) Sub-cellular localisation of GmLUXb and GmLUXc proteins in onion epidermal cells. Onion epidermal peels were bombarded with constructs 35S-GFP alone or 35S-GFP fused with either GmLUXb or GmLUXc sequences as described in methods and materials section. When GFP was expressed alone, the green fluorescence was dispersed throughout the cell, while green fluorescence from GFP-LUX fusion proteins were localized in nuclei. GFP: green fluorescence, DAPI: stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), BF: under bright field. (C) Homologues of soybean LUX gene show tissue-specific expression. Expression of GmLUXa, GmLUXb, and GmLUXc in shoot apical meristem (SAM), flowers, green pods, leaves, nodules, root, and root tips. (D) Homologues of soybean LUX gene show diurnal and circadian rhythms (i) Diurnal rhythms of GmLUXb and GmLUXc under long-day (LD; 16 h light, 8 h dark) or short-day (SD; 10 h light, 14 h dark) conditions. (ii) Circadian rhythms of GmLUXb and GmLUXc under constant light (LL; 12 h light:12 h dark then continuous light). All plants were three weeks old at the time of sampling. Data are mean ± SE for n = three biological replicates, each consisting of pooled material from two plants. Day and night periods are depicted above the graph by open and closed bars, respectively.