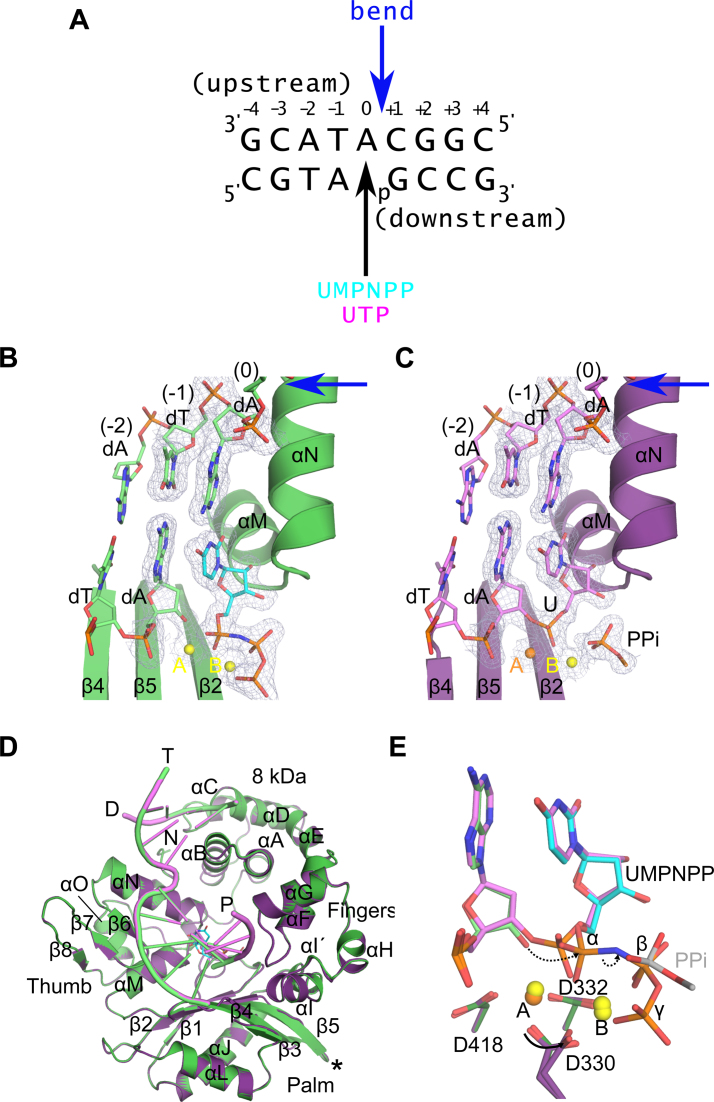
Figure 1.
Structural evaluation of ribonucleotide incorporation by wildtype hPol μΔ2. Wildtype hPol μΔ2 was crystallized in complex with a 1-nt gapped DNA substrate and an incoming dUMPNPP (A). A pre-catalytic ternary complex (B, green) was obtained by soaking with nonhydrolyzable UMPNPP (cyan), and a post-catalytic nicked complex (C, purple) was obtained by soaking with hydrolyzable UTP. Structures of the active sites of each complex are shown, with the protein drawn in cartoon and the DNA substrate in stick. Protein secondary structural elements are marked, with β-strands numbered, and α-helices labeled alphabetically. Magnesium (yellow) and manganese (orange) ions are drawn as spheres. 2Fo – Fc electron density (contoured at 1σ) is shown for the DNA substrate near the catalytic center of each complex. The location of the 90° bend in the DNA template strand is indicated by a blue arrow. The partially disordered pyrophosphate leaving group is shown in stick. (D) Global superpositions of the pre- (green) and post-catalytic (purple) complexes. The location of the Loop2 truncation is shown as a black asterisk. (E) Detailed comparison of the wildtype hPol μΔ2 active center in the pre- (protein in dark green; primer terminus in light green; UMPNPP in cyan), and post-catalytic (protein in purple; DNA in lavender) complexes. Dashed arrows indicate movement of electrons during progression of the reaction. All structural figures were generated using PyMOL (Schrödinger).