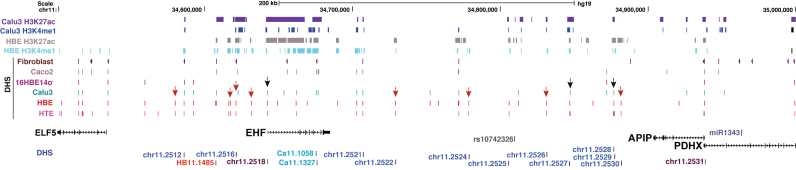
Figure 1.
The chromatin landscape of the 11p13 CF modifier region. UCSC genome browser graphic shows a 500 kb window surrounding the highest P-value SNPs at 11p13 between EHF and APIP, with the most significant GWAS I+II SNP (rs10742326) shown in grey. ChIP-seq for the active histone marks H3K27ac and H3K4me1 in Calu3 (20) and primary HBE (28) cells are shown at the top. Below are DNase-seq data showing open chromatin regions as DNase hypersensitive sites (DHS) in fibroblast (GM03348, http://www.ncbi.nlm.nih.gov/geo/GSM1008563 (33)), Caco2 (28), 16HBE14o-, Calu3 (20), HBE (28) and HTE (29) cells. The location of genes in the region is shown underneath, with cloned fragments assayed in reporter genes and key genomic features relevant to experiments shown below.