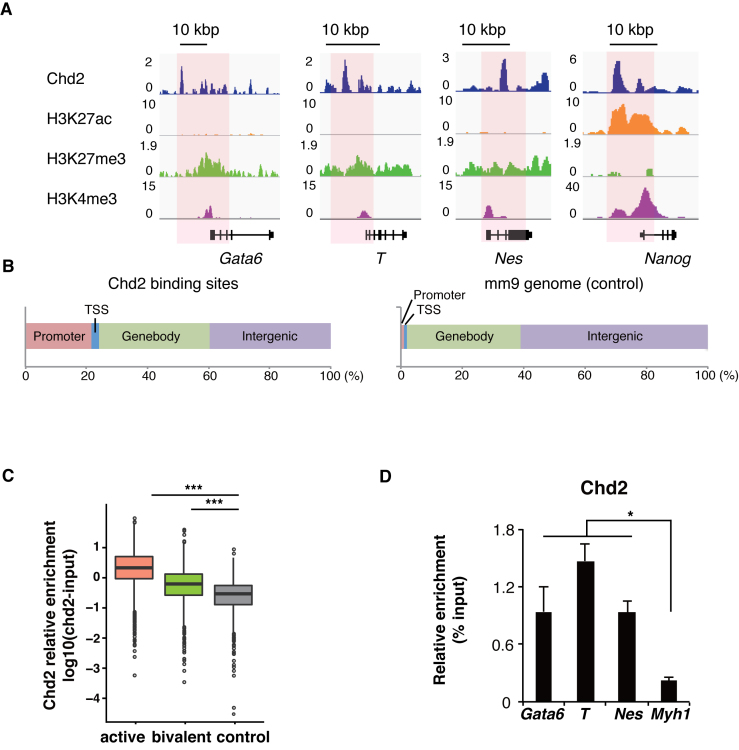
Figure 3.
Chd2-dependent developmental genes are in the bivalent state in undifferentiated mESCs. ChIP-seq data of histone modifications were obtained from WT ESCs and analyzed with the ENCODE dataset of Chd2 in mESCs. (A) Genome browser tracks representing Chd2, H3K27ac, H3K27me3 and H3K4me3 occupancy around bivalent genes (Gata6, T and Nes) and an active gene (Nanog). Read counts were normalized to the total number of reads for each dataset. (B) Left bar chart shows the distribution of Chd2-binding regions in the mm9 genome annotated by HOMER (43). Right bar chart shows the reference proportion of the mm9 genome. (C) ChIP-seq enrichment of Chd2 mapped around TSSs (±1 kbp) in each gene subset. (D) ChIP-qPCR assay using anti-Chd2 antibodies in EBs derived from WT ESCs. Both bivalent genes (Gata6, T and Nes) and a silent gene (Myh1) were analyzed. Recovery efficiency (mean ± standard deviation of three independent experiments) is expressed as enrichment relative to the input. *P < 0.05.