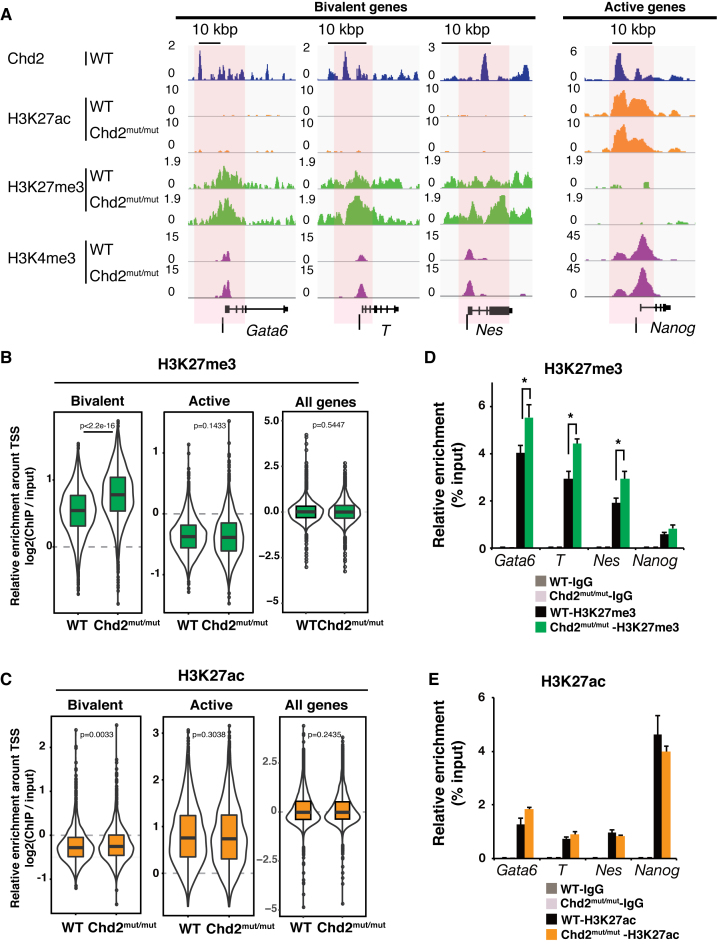
Figure 4.
Chd2 prevents suppressive chromatin formation at Chd2-dependent developmental genes. ChIP-seq data of histone modifications were compared between WT and Chd2mut/mut ESCs. (A) Genome browser representing H3K27ac, H3K27me3 and H3K4me3 occupancy around bivalent state genes (Gata6, T and Nes) and an active state gene (Nanog) in WT and Chd2mut/mut ESCs. Black bars indicate the regions selected for ChIP-qPCR. (B) Box plots show H3K27me3 and (C), H3K27ac ChIP-seq signal intensities for the bivalent, active, and all genes at the TSS ± 5 kbp. The Wilcoxon test was used for statistical analysis. (D) ChIP-qPCR assay using anti-H3K27me3 and (E) anti-H3K27ac antibodies. Both bivalent genes (Gata6, T and Nes) and an active gene (Nanog) were analyzed in WT and Chd2mut/mut ESCs. Recovery efficiency (mean ± standard deviation of three independent experiments) is expressed as enrichment relative to the input. *P < 0.05.