Abstract
We have undertaken a systematic structural study of Thermus thermophilus Argonaute (TtAgo) ternary complexes containing single-base bulges positioned either within the seed segment of the guide or target strands and at the cleavage site. Our studies establish that single-base bulges 7T8, 5A6 and 4A5 on the guide strand are stacked-into the duplex, with conformational changes localized to the bulge site, thereby having minimal impact on the cleavage site. By contrast, single-base bulges 6’U7’ and 6’A7’ on the target strand are looped-out of the duplex, with the resulting conformational transitions shifting the cleavable phosphate by one step. We observe a stable alignment for the looped-out 6’N7’ bulge base, which stacks on the unpaired first base of the guide strand, with the looped-out alignment facilitated by weakened Watson–Crick and reversed non-canonical flanking pairs. These structural studies are complemented by cleavage assays that independently monitor the impact of bulges on TtAgo-mediated cleavage reaction.
INTRODUCTION
Much effort has been directed toward a mechanistic understanding of the role of Argonaute (Ago) proteins in RNA silencing (1–6). Ago proteins accommodate guide strands that function as templates for pairing and subsequent cleavage of complementary target strands. Insights into Ago-mediated nucleation, propagation and cleavage of target strands has emerged from structural studies of Thermus thermophilus Ago (TtAgo) binary complexes with 5′-phosphorylated DNA guide strands (7) and ternary complexes with added RNA (8) and DNA (9) target strands. Recent functional studies have demonstrated that TtAgo acts as a barrier for the uptake and propagation of foreign DNA, thereby functioning in host defense by a DNA-guided DNA interference pathway (10). Related insights into host defense have independently emerged from functional studies undertaken on Rhodobacter sphaeroides Ago ternary complexes (11). These studies on prokaryotic Ago complexes have been extended to binary complexes of eukaryotic Agos with bound RNA guide strands from budding yeast (12) and humans (13,14) and on ternary complexes with added RNA targets on human Ago2 (15). The structural research has highlighted the role of the MID and PIWI domains in recognizing the 5′-phosphate (16,17) and the PAZ domain in recognizing the 3′-ends (18,19) of the guide respectively, and the conserved acidic residues positioned within the RNase H fold of the PIWI domain (20–22) in catalytic cleavage activity. In addition, conformational transitions have been identified on proceeding from the structure of Ago in the free state, to its binary complex with guide strand and ternary complex with added target strand (7,8,15). Several reviews have highlighted the structural basis underlying the role of prokaryotic and eukaryotic Ago proteins in RNA silencing (23–25).
Mismatches and bulges (reviewed by Hermann (26)) are a common feature of miRNA–target–RNA interactions (reviewed by Bartel (27)) and it has remained a challenge as to how these helical imperfections are accommodated within the nucleic acid-binding channel of the Ago scaffold and how the resulting structural distortions impact on the positioning of the scissile phosphate relative to the catalytic acidic residues lining the binding pocket of the RNase H-like PIWI domain fold. Towards this goal, we have generated and solved crystal structures of TtAgo ternary complexes containing single base bulges positioned within the seed segment on both guide and target strands and positioned within the cleavage site on the target strand. These structural studies have allowed us to differentiate between stacked-in and looped-out alignments of the bulge base, and in addition have provided insights into potential long-range perturbations associated with single bulge base sites. Most importantly, our studies highlight the identification of a stable conformation involving a looped-out bulge between positions 6′ and 7′ on the target strand.
These structural studies have been complemented by cleavage assays undertaken at elevated temperatures for optimal cleavage that provide an independent measure of the ability of the TtAgo scaffold to accommodate bulges on the guide and target strands within the seed segment and their impact on site-specific cleavage of the target strand.
MATERIALS AND METHODS
Crystallization and data collection
Wild-type and mutant T. thermophilus Ago were prepared as described previously (7). Oligodeoxynucleotides were purchased from Invitrogen. RNA oligonucleotides were purchased from Dharmacon. For crystallization, T. thermophilus Ago was mixed with 5′-phosphorylated 21-mer guide DNA at 1:1.2 molar ratio, followed by addition of target DNAs or RNAs at a 1.0 molar ratio to the binary mixture, to form the ternary complex. All crystals were grown at 35°C.
Diffraction data were collected on beamline NE-CAT ID-24C at the Advanced Photon Source (APS), Argonne National Laboratory and beamline X-29 at the Brookhaven National Laboratory. All data sets were integrated and scaled with the HKL2000 suite (28) and data processing statistics are summarized in Supplementary Tables S1 and S2.
Crystals of catalytic N546 mutant Ago complexed with 7T8 bulge-containing guide DNA and 16-nt target DNA were grown from 3.0 M NaAc·3H2O, 100 mM Bis-Tris propane, pH 7.0–7.2. Crystals of catalytic N546 mutant Ago complexed with 5A6 bulge-containing guide DNA and 16-nt target DNA were grown from 2.8 M Na-acetate, pH 7.0, 0.1 M Tris–Cl, pH 8.0 and 0.15 M glycine. Crystal of wild-type Ago complexed with 4A5 bulge-containing guide DNA and 16-nt target DNA were grown from 2.6 M Na-acetate, pH 7.0, 0.1 M Tris–Cl, pH 7.4 and 0.2 M glycine.
Crystals of catalytic N546 mutant Ago complexed with 6’U7’ bulge-containing target RNA were grown from 1.0 M (NH4)2SO4, 0.1 M KCl, 10 mM MgCl2, 50 mM Bis–Tris pH 6.5 by hanging-drop method. Crystals of catalytic N546 mutant Ago complexed with 6’A7’ bulge-containing target RNA were grown from 2.0 M Na-formate and 0.1 M Tris–Cl pH 7.2–7.5. Crystals of catalytic N546 mutant Ago complexed with 9’U10’ bulge-containing target RNA were grown from 3.1 M NH4-acetate, 0.1 M Bis–Tris propane, pH 7.3.
Structure determination and refinement
The structures of the complexes were solved by molecular replacement with the program PHASER (29). The domains of the Ago-21-mer guide DNA binary complex structure (7) without the linkers were used as search models. Model building was done using COOT (30), and refinement was done with CNS (31) and PHENIX (32). The final Figures were created with Pymol (http://pymol.sourceforget.net/). The refinement statistics for all the Ago mutants and wild-type complexes are summarized in Supplementary Tables S1 and S2.
Oligonucleotides
DNA guide let-7, 5′-TGAGGTAGTAGGTTGTATAGT; 4A5, 5′-TGAGAGTAGTAGGTTGTATAGT; 4C5, 5′-TGAGCGTAGTAGGTTGTATAGT; 4G5, 5′-TGAGGGTAGTAGGTTGTATAGT; 4T5, 5′-TGAGTGTAGTAGGTTGTATAGT; 5A6, 5′-TGAGGATAGTAGGTTGTATAGT; 5C6, 5′-TGAGGCTAGTAGGTTGTATAGT; 5G6, 5′-TGAGGGTAGTAGGTTGTATAGT; 5T6, 5′-TGAGGTTAGTAGGTTGTATAGT; 7A8, 5′-TGAGGTAAGTAGGTTGTATAGT; 7C8, 5′-TGAGGTACGTAGGTTGTATAGT; 7G8, 5′-TGAGGTAGGTAGGTTGTATAGT; 7T8, 5′-TGAGGTATGTAGGTTGTATAGT. DNA target let-7, 5′-TATACAACCTACTACCTCG; 6΄A7΄, 5΄-TATACAACCTACTAACCTCG; 6΄C7΄, 5΄-TATACAACCTACTCACCTCG; 6΄G7΄, 5΄-TATACAACCTACTGACCTCG; 6΄T7΄, 5΄-TATACAACCTACTTACCTCG; 7΄A8΄, 5′-TATACAACCTACATACCTCG; 7΄C8΄, 5′-TATACAACCTACCTACCTCG; 7΄G8΄, 5′-TATACAACCTACGTACCTCG; 7΄T8΄, 5′-TATACAACCTACTTACCTCG; 9’T10’, 5′-TATACAACCGTTCTACCTCG. RNA target let-7, 5′-UAUACAACCUACUACCUCG; 6’U7’, 5′-UAUACAACCUACAUACCUCG; 6’A7’, 5′-UAUACAACCUACUAACCUCG; 9’U10’, 5′-UAUACAACCGUUCUACCUCG.
Guide and target oligonucleotides were 5′-phosphorylated using T4 polynucleotide kinase and non-radioactive ATP or γ-32P-ATP, respectively, followed by size exclusion chromatography over G25 columns (GE Healthcare illustra Microspin) or purification using a 15% denaturing polyacrylamide gel, respectively.
In vitro cleavage assays of TtAgo
First, 5 μM recombinant TtAgo was loaded with 0.5 μM guide DNA in 10 mM HEPES-KOH, pH 7.5, 100 mM NaCl, 5 mM MgCl2, for 30 min at 55°C in a volume of 10 μl per reaction. Then, 0.5 μM 5′-32P-radiolabeled DNA or RNA target was added, and incubation continued at 75°C for indicated times. Reactions were terminated by addition of an equal volume of stop solution, containing 8 M urea, 50 mM EDTA, and 0.3 mg/ml bromophenol blue. Products were heated for 5 min at 95°C, placed on ice for 2 min, resolved on 15% denaturing polyacrylamide gels, and visualized by phosphorimaging. Signal quantitation was performed with ImageJ software (http://imagej.nih.gov/ij). Hydrolysis of input RNA was performed in 0.1 M NaOH at 95°C for 1 min. Hydrolysis products carry a 2′,3′-cyclic phosphate that also convert to 2′ or 3′ monophosphates and run faster than the respective TtAgo cleavage products that carry a 3′ hydroxyl.
RESULTS
We have undertaken a combined structural and enzymatic cleavage investigation to evaluate the impact of single base bulges positioned within the seed segment on the guide and target strands and at the cleavage site, both on the alignment and perturbation of the guide-target duplex within the TtAgo scaffold. Towards this end, we investigated the crystallization propensity of TtAgo ternary complexes containing single base pyrimidine and purine bulges and report below on several systems that yielded diffraction quality crystals of bulge-containing TtAgo ternary complexes. The study initially outlines three examples of bulges at three different positions within the seed segment of the guide strand, and is followed by two examples of bulges within the seed segment of the target strand. Finally, an example is presented of a bulge positioned within the cleavage site of the target strand. The structural studies on ternary complexes reported below were primarily undertaken on the N546 catalytic mutant of TtAgo (to prevent target strand cleavage) bound by a 5′-phosphorylated 21-mer DNA guide (22-mer in case of a bulged base) and complementary 19-mer DNA and RNA targets (20-mer in case of a bulged base).
Both structural and functional studies have established that TtAgo containing a 5′-phosphorylated DNA guide can cleave both RNA and DNA targets (8–10). We have previously undertaken structural studies on TtAgo ternary complexes on both DNA (9) and RNA (8) targets. Our initial structural studies with bulge bases positioned within the seed segment and the cleavage site on the target strand were undertaken with RNA targets. Subsequent studies on bulge bases within the seed segment on the guide strand were undertaken with DNA targets, given the advantage of DNA targets being more stable than their RNA counterparts. The TtAgo-mediated cleavage assays were undertaken on the same bulge-containing guide-target duplexes for which crystal structures were available and performed at 75°C as required for optimal activity of this thermophilic Ago system.
Structures of single base bulges within seed segment of the guide strand
Structural studies of single base bulges within the seed segment of the guide strand were undertaken on TtAgo ternary complexes containing DNA guide and DNA target strands. These include bulges between positions 7 and 8 on the guide strand (designated 7T8 bulge), as well as 4A5 and 5A6 bulge sites.
7T8 bulge
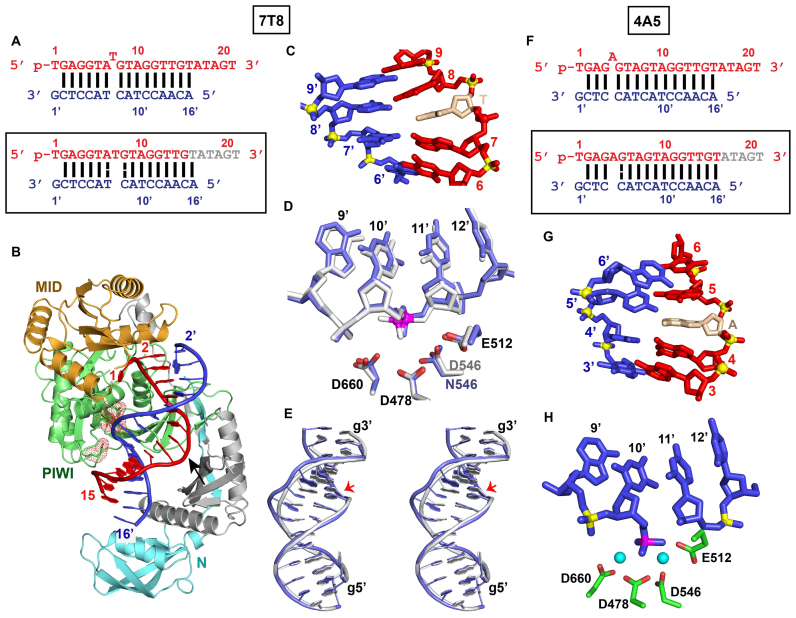
We have solved the 2.9 Å crystal structure of the TtAgo (D546N catalytic mutant) ternary complex containing a bulged thymine between positions 7 and 8 on the guide strand (designated 7T8 bulge; Figure 1A; x-ray statistics listed in Supplementary Table S1). The 2Fo-Fc electron densities for the entire duplex and for the segment centered about the stacked-in T bulge site in the ternary complex are shown in Supplementary Figure S1A (in stereo) and 1, respectively. It should be noted that the PAZ domain is disordered and could not be traced in this complex (Figure 1B). The key observation is that the T bulge stacks into the duplex between A7 and G8 on the guide strand (Figure 1C and Supplementary Figure S1B), where it positioned between weakened flanking Watson–Crick A7-T7’ and G8-C8’ base pairs (Supplementary Figure S1C; see schematic in Figure 1A, lower panel; weakened base pairs labeled by dashed lines).
Figure 1.
Crystal Structure of TtAgo Bound to 5′-phosphorylated 22-nt Guide DNA and 19-nt Target DNA Containing a 7T8 (TtAgo D546N Catalytic Mutant) and 4A5 (TtAgo Wild-type) Bulges Positioned Within the Seed Segment on the Guide Strand. (A) Sequence of the guide DNA–target DNA duplex (top panel), with the actual alignment of the bulge in the crystal structure of the ternary complex (bottom boxed panel), where a thymine stacks into the duplex between positions 7 and 8 of the guide strand. The traceable segments of the nucleotides of the guide DNA and target DNA in the structure of the ternary complex are shown in red and blue, respectively. The dashed lines show weakened Watson–Crick pairs. (B) 2.9 Å structure of TtAgo (N546 catalytic mutant) bound to 5′-phosphorylated 22-nt guide DNA (in red) and 19-nt target DNA (in blue) containing a 7T8 bulge positioned on the guide strand within the seed segment. There is one molecule of the complex in the asymmetric unit. The PAZ domain is disordered and the 3′-end of the guide strand cannot be monitored in the complex. (C) A stick representation of the bulge site and two flanking base pairs, with the stacked-in thymine highlighted in biscuit color in the 7T8 bulge-containing ternary complex. (D) The positioning of the DNA target strand of the control containing no bulge (in silver) and in the 7T8 bulge (in blue) relative to the catalytic residues (D478, D660, E512 and D546N mutant) of the RNase H fold of the PIWI domain in the TtAgo ternary complex. The catalytic residues are equidistant from the phosphate linking the 10’–11’ step (colored in magenta) in the control (in silver) and 7T8 bulge (in blue)-containing Ago ternary complexes. (E) Superposition of the guide-target duplex containing no bulge (in silver) and 7T8 bulge (in blue) in Ago ternary complexes. The guide strand is labeled g and the 10’–11’ phosphate at the cleavage site on the target strand is indicated by a red arrow. (F) Sequence of the guide DNA–target DNA duplex (top panel), with the actual alignment of the bulge in the crystal structure of the ternary complex (bottom boxed panel), where a adenine stacks into the duplex between positions 4 and 5 of the guide strand. Wild-type TtAgo was used to generate crystals of this complex. (G) A stick representation of the bulge site and two flanking base pairs, with the stacked- in adenine highlighted in biscuit color in the 3.1 Å structure of the 4A5 bulge-containing ternary complex. There is one molecule of the complex in the asymmetric unit and the 3′-end of the guide strand is inserted into the PAZ pocket of an adjacent molecule in the crystal lattice (not shown). (H) The positioning of the DNA target strand of the 4A5 bulge (in blue) relative to the catalytic residues (D478, D660, E512 and D546) of the RNase H fold of the PIWI domain in the wild-type TtAgo ternary complex. Note that the backbone has cleaved at the 10’–11’ step in the target strand and that a pair of Mg2+ cations were identified at the cleavage site in the wild-type TtAgo ternary complex.
We have compared the relative positioning of the cleavable phosphate and the catalytic tetrad in the 7T8 bulge (in blue) relative to control lacking a bulge (in silver) at the Ago ternary complex level (Figure 1D). Notably, there is minimal distortion at the 10’–11’ cleavage site on the target strand (Figure 1D) as a result of a stacked-in T bulge between bases 7 and 8 of the guide strand in the ternary Ago complex. In addition, we have superposed the entire guide-target duplex of the 7T8 bulge (in blue) relative to control lacking a bulge (in silver) and minimal differences were observed (cleavage site indicated by red arrow) as shown in a stereo view in Figure 1E. Similarly, we have compared surface views of Ago in non-bulge containing control and 7T8 bulge-containing ternary TtAgo complexes in Supplementary Figure S1d and e, respectively, and observed minimal differences aside from the disordered PAZ domain in the 7TA bulge-containing ternary complex.
4A5 bulge
We next solved the 3.1 Å structure of a TtAgo ternary complex containing an adenine bulge between positions 4 and 5 (4A5 bulge) within the seed segment of the guide strand. Attempts to obtain diffraction quality crystals for the ternary complex using the D546N catalytic mutant of TtAgo were unsuccessful, and hence the structure was solved for the ternary complex using wild-type TtAgo (Figure 1F, top panel; X-ray statistics in Supplementary Table S1). The overall structure of the complex is shown in Supplementary Figure S2A, with the PAZ domain traceable in this complex. The bulged adenine stacks into the duplex for the 4A5 bulge (Figure 1G and Supplementary Figure S2B). We observe weakening of G5-C5’ Watson–Crick base pair flanking the inserted adenine (Supplementary Figure S2C; see schematic in Figure 1F, boxed bottom panel) and a cleaved phosphodiester bond at the 10’–11’ step for the 4A5 bulge-containing TtAgo (wild-type) ternary complex (Figure 1H and Supplementary Figure S2D). We also observe a pair of Mg2+ ions at the cleavage site (Figure 1H and Supplementary Figure S2D).
5A6 bulge
We also solved the 2.8 Å structure of a TtAgo (D546N catalytic mutant) ternary complex containing an adenine bulge between positions 5 and 6 (5A6 bulge) within the seed segment of the guide strand (Figure 2A, top panel; X-ray statistics in Supplementary Table S1). The overall structure of the complex is shown in Supplementary Figure S2E, with the PAZ domain also traceable in this complex. The bulged adenine stacks into the duplex for the 5A6 bulge (Figure 2B and Supplementary Figure S2F), with minimal impact in the positioning of the cleavable phosphate at position 10’–11’ on the target strand (Figure 2C). We observe weakening of G5-C5’ Watson–Crick base pair flanking the inserted adenine (Supplementary Figure S2G; see schematic in Figure 2A, boxed bottom panel) and an intact phosphodiester bond at the 10’-11’ step for the 5A6 bulge-containing TtAgo (D546N catalytic mutant) ternary complex (Figure 2C and Supplementary 2H).
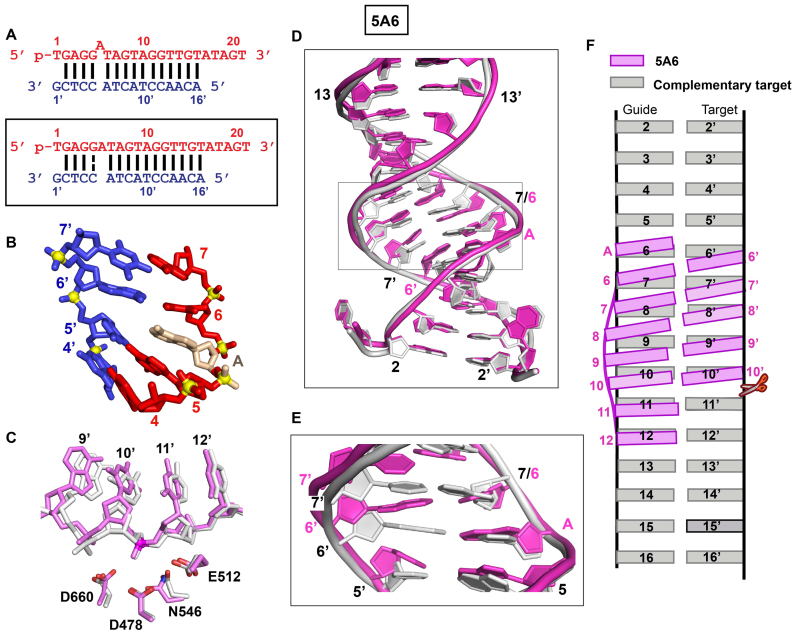
Figure 2.
Crystal structures of TtAgo (TtAgo D546N catalytic mutant) bound to 5′-phosphorylated 22-nt guide DNA and 19-nt target DNA containing a 5A6 bulge positioned on the guide strand within the seed segment and conformational adjustments on proceeding from control to 5A6 bulge-containing ternary complexes. (A) Sequence of the guide DNA–target DNA duplex (top panel), with the actual alignment of the bulge in the crystal structure of the ternary complex (bottom boxed panel), where an adenine stacks into the duplex between positions 5 and 6 of the guide strand. (B) A stick representation of the bulge site and two flanking base pairs, with the stacked- in adenine highlighted in biscuit color in the 2.8 Å structure of the 5A6 bulge-containing ternary complex. There are two molecules of the complex in the asymmetric unit and the 3′-end of the guide strand is inserted into the PAZ pocket of an adjacent molecule in the crystal lattice (not shown). (C) The positioning of the DNA target strand of the control containing no bulge (in silver) and in the 5A6 bulge (in magenta) relative to the catalytic residues (D478, D660, E512 and D546N mutant) of the RNase H fold of the PIWI domain in the TtAgo ternary complexes. The catalytic residues are equidistant from the phosphate linking the 10’–11’ step (colored in red) in the control (in silver) and 5A6 bulge (in magenta)-containing Ago ternary complexes. (D, E) Superposition of the seed and cleavage site segments of the guide-target duplex in the no-bulge control (in silver) and 5A6 bulge-containing (in magenta) Ago ternary complexes. The segment spans 1–1’ to 14–14’ in panel D and spans 5–5′ to 8–8’ in panel E. (F) Schematic emphasizing base tilting of the guide strand between 5–5′ and 11–11’ pairs in the duplex of the 5A6 bulge-containing Ago ternary complex (in magenta), relative to the duplex of the no-bulge control ternary complex (in silver).
We have superposed the control duplex lacking the bulge (in silver, Figure 2D and E) with the duplex containing the 5A6 bulge (in magenta, Figure 2D and E) and note that stacking-in of the adenine in the 5A6 bulge results in tilting of the base pairs from 6–6’ to 12–12’ in the 5A6 bulge-containing Ago ternary complex (in magenta, Figure 2F), with minimal change in the position of the target strand at the 10’–11’ cleavage site.
Structure of single base bulges within seed segment of the target strand
Structural studies of single base bulges within the seed segment of the target strand were undertaken on TtAgo ternary complexes containing DNA guide and RNA target strands. These include bulges containing an adenine between positions 7’ and 8’, as well as between 6’ and 7’ on the target strand.
6’U7’ bulge
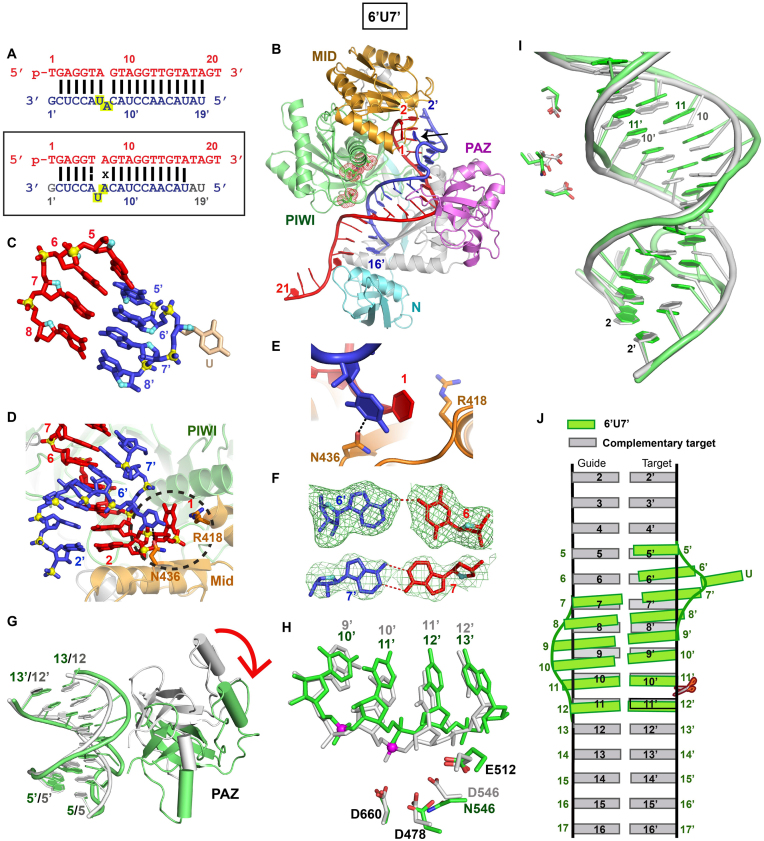
Our initial studies focused on a bulge sequence that was designed to contain a bulged adenine between positions 7’ and 8’ on the target RNA strand (Figure 3A, top panel), but following structural analysis was found instead to contain a bulged uracil between positions 6’ and 7’ on the target strand (Figure 3A, boxed bottom panel). Henceforth, this bulge-containing sequence will be labeled 6’U7’ bulge. We have solved the 2.8 Å crystal structure of the TtAgo (D546N catalytic mutant) ternary complex containing a 6’U7’ bulge. The overall structure of the complex is shown in Figure 3B (X-ray statistics listed in Supplementary Table S2), with the packing arrangement of two ternary complexes in the crystal lattice shown in Supplementary Figure S3A. We were able to trace the DNA guide strand along its entire length from positions 1 to 21 (Figure 3B), with its 3′-end anchored in the PAZ domain of the second adjacent Ago molecule in the crystal lattice (Supplementary Figure S3A). The bound RNA target can be traced from positions 2’ to 16’ in the ternary complex (Figure 3B and Supplementary Figure S3B), and we can also confidently trace the electron density in the vicinity of the bulge site (Supplementary Figure S3C). To our surprise, it is the uracil that is 3′ to the putative bulged adenine on the target strand, that is looped-out of the duplex (Figure 3C and Supplementary Figure S3C; schematic in Figure 1A, bottom boxed panel) and stacks over the unpaired first base at the 5′-end of the guide strand (Figure 3d and e), and is further anchored in place through a hydrogen bond with an Asn side chain (Figure 3E).
Figure 3.
Crystal structure of TtAgo (D546N catalytic mutant) bound to 5′-phosphorylated 21-nt guide DNA and 20-nt target RNA containing a 6’U7’ bulge positioned on the target strand within the seed segment and conformational adjustments on proceeding from control to 6’U7’ bulge-containing ternary complexes. (A) Sequence of the intended guide DNA–target RNA duplex (top panel), with the actual alignment of the bulge in the crystal structure of the ternary complex (bottom boxed panel), where an adjacent uracil loops out of the duplex between positions 6’ and 7’ of the target strand. The traceable segments of the nucleotides of the guide DNA and target RNA in the structure of the ternary complex are shown in red and blue, respectively. The dashed line for the T6–A6’ pair represents a weakened Watson–Crick pair stabilized by one hydrogen bond, while x shown for the A7•A7’ pair represents a reversed non-canonical pairing alignment. (B) 2.8 Å structure of TtAgo (D546N catalytic mutant) bound to 5′-phosphorylated 21-nt guide DNA (in red) and 20-nt target RNA (in blue) containing a 6’U7’ bulge positioned on the target strand within the seed segment. The black arrow points to the looped out bulge base. There is one molecule of the complex in the asymmetric unit and the 3′-end of the guide strand is inserted into the PAZ pocket of an adjacent molecule in the crystal lattice (see Supplementary Figure S3A). (C) A stick representation of the bulge site and two flanking base pairs on either side, with the looped out uracil highlighted in biscuit color in the 6’U7’ bulge-containing ternary complex. Note the opposing directionalities of the sugar ring oxygens (in cyan) on either side of the bulge site. (D) An overall view highlighting the looped-out uracil of the target RNA and its stacking on sheared-apart base 1 of the guide DNA in the 6’U7’ bulge-containing ternary complex. (E) Stacking of the looped our uracil with unpaired first base of the guide strand and hydrogen bonding with the side chain of Asn436 in the 6’U7’ bulge-containing ternary complex. (F) An omit map (3σ) identifying pairing alignment for the weakened Watson–Crick A6•U6’ pair (top panel) and the reversed A7•A7’ pair (bottom panel) flanking the bulged looped out uracil in the 6’U7’ bulge-containing ternary complex. (G) Superposition of the PAZ domain and guide-target duplex containing no bulge (in silver) and 6’U7’ bulge (in green) in Ago ternary complexes. A red arrow indicates the large conformational transition observed for the PAZ domain on proceeding from the no bulge (in silver) to 6’U7’ bulge (in green) ternary complexes. (H) The positioning of the RNA target strand in the no bulge-containing control (in silver) and in the 6’U7’ bulge-containing (in green) Ago ternay complexes relative to the catalytic residues of the RNase H fold of the PIWI domain in the complex. The catalytic residues are distant from the phosphate linking the 10’–11’ step (colored in magenta) and closer to the phosphate linking the 11’–12’ step in the 6’U7’ bulge-containing Ago ternary complex (in green). (I) Superposition of the seed and cleavage site segments of the guide-target duplex in the no-bulge control (in silver) and 6’U7’ bulge-containing (in green) Ago ternary complexes. (J) Schematic emphasizing base displacement of the target strand between 5–5′ and 11–11’ pairs in the duplex of the 6’U7’ bulge-containing Ago ternary complex (in green), relative to the duplex of the no-bulge control ternary complex (in silver).
The looped-out uracil is flanked by a weakened Watson–Crick A-U (Figure 3F, top panel; designated by a dashed line in Figure 3A, boxed bottom panel) and reversed Watson–Crick edge-aligned non-canonical A•A (Figure 3F, bottom panel; designated by an x in Figure 3A, boxed bottom panel) pairs, with the localized distortion within this segment of the seed region propagated to the cleavage site. Conformational perturbations are observed for both the guide-target duplex and the PAZ domain on proceeding from Ago ternary complex with control RNA target (in silver, Figure 3G) to that with 6’U7’ bulge-containing RNA target (in green, Figure 3G). As a result the 6’U7’ bulge-containing duplex exhibits a wider groove and a compression-related shift by one base pair within the guide (5–13)•target (5′-13’) segment of the duplex (Figure 3G). These changes are also accompanied by a conformational transition within the PAZ domain (see red arrow, Figure 3G; see also surface views of Ago in non-bulge containing control and 6’U7’ bulge-containing ternary TtAgo complexes in Supplementary Figure S3d and S3e, respectively). Thus, the phosphate at the 10’–11’ step in the 6’U7’ bulge-containing duplex is no longer positioned opposite the catalytic tetrad (D478, D660, E512 and N546 mutant), resulting in the cleavage site being shifted by one phosphate towards the 11’–12’ position (Figure 3H). This shift in the cleavage site can also be visualized following superposition of the control (in silver) and 6’U7’-bulge (in green)-containing Ago ternary complexes, as shown schematically in Figure 3J.
These structural results provide insights regarding the specificity of target recognition and illustrate how what was assumed to be a 7’A8’ bulge in the target strand (Figure 3A, upper panel) turned out instead to be a 6’U7’ looped-out U bulge (Figure 3A, lower boxed panel), that was accommodated by long range perturbations in the positioning of the guide-target duplex (Figure 3H–J) within the nucleic acid-binding channel of the Ago scaffold.
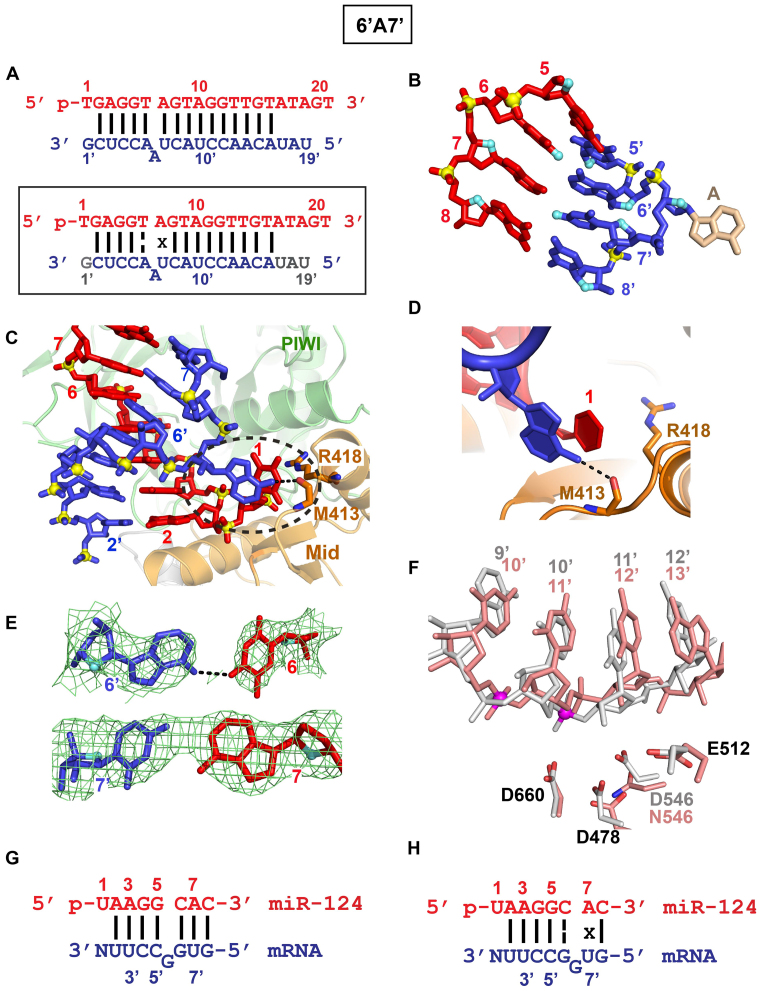
6’A7’ bulge
We have also solved the 3.2 Å crystal structure of the TtAgo ternary complex containing a bulged adenine between positions 6’ and 7’ on the target strand (designated 6’A7’ bulge; Figure 4A, top panel). The overall structure (Supplementary Figure S3F), packing arrangement and crystallographic statistics (Supplementary Table S2) are the same for the structures of the ternary complexes containing the 6’U7’ bulge (Figure 3A, bottom boxed panel) presented above and the 6’A7’ bulge (Figure 4A, bottom boxed panel). The extra adenine is looped-out of the duplex (Figure 4B and Supplementary Figure S3G) and stacks over the unpaired first base at the 5′-end of the guide strand (Figure 4C, D and Supplementary Figure S3G). The looped out adenine is hydrogen bonded to the side chain of a Met 413 (Figure 4D). The T6–A6’ pair on one side of the 6’A7’ bulge adopts the weakened Watson–Crick alignment stabilized by one hydrogen bond (Figure 4E, top panel), while the A7•U7’ pair on the other side adopts a reversed A•U alignment stabilized by one hydrogen bond (Figure 4E, bottom panel), resulting in the sequence alignment shown in Figure 4A, boxed bottom panel (dashed line indicates weakened Watson–Crick pairing and x indicates reversed pairing). Further, the localized distortion associated with the looped-out 6’A7’ adenine bulge on the target strand in the seed segment is propagated to the cleavage site, such that the cleavage site is once again shifted by one phosphate towards the 11’–12’ position (Figure 4F; control in silver and 6’A7’ bulge in salmon).
Figure 4.
Crystal structure of TtAgo (D546N catalytic mutant) bound to 5′-phosphorylated 21-nt guide DNA and 20-nt target RNA containing a 6’A7’ bulge positioned within the seed segment on the RNA target strand and alternate alignments of guanine bulge associated with pairing of miR-124 guide and its complementary mRNA target. (A) Sequence of the guide DNA-target RNA duplex (top panel), with the actual alignment of the bulge in the crystal structure of the ternary complex (bottom boxed panel), where the adenine loops-out of the duplex between positions 6’ and 7’ of the target strand. The traceable segments of the nucleotides of the guide DNA and target RNA in the structure of the ternary complex are shown in red and blue, respectively. The dashed line shown for T6–A6’ represents a weakened Watson–Crick pair stabilized by one hydrogen bond. The x shown for A7•U7’ represents a reversed non-canonical pairing alignment. (B) A stick representation of the bulge site and two flanking base pairs, with the looped out adenine highlighted in biscuit color in the 6’A7’ bulge-containing ternary complex. Note the opposing directionalities of the sugar ring oxygens (in cyan) on either side of the bulge site. (C) An overall view highlighting the looped-out adenine of the target RNA and its stacking on the unpaired first base of the guide DNA in the 6’A7’ bulge-containing ternary complex. (D) Stacking of the looped our adenine with sheared-apart base 1 of the guide strand and hydrogen bonding with the side chain of Met413 in the 6’A7’ bulge-containing ternary complex. (E) Omit maps (3σ) identifying pairing alignment of the weakened Watson–Crick T6•A6’ pair stabilized by one hydrogen bond (denoted by dashed line in panel A, boxed bottom panel) and reversed A7•U7’ pair (denoted by x in panel A, boxed bottom panel) flanking the looped-out adenine in the 6’A7’ bulge-containing ternary complex. (F) The positioning of the RNA target strand of the control Ago ternary complex containing no bulge (in silver) and in the 6’A7’ bulge (in salmon) relative to the catalytic residues (D478, D660, E512 and D546N mutant) of the RNase H fold of the PIWI domain in the complex. The catalytic residues are distant from the phosphate linking the 10’–11’ step (colored in magenta) and closer to the phosphate linking the 11’–12’ step in the 6’A7’ bulge (in salmon)-containing Ago ternary complex. (G) The guanine bulge is positioned at 5’G6’ on the target strand following the analysis by Chi et al. (2012). The dots represent non-canonical pairs. (H) An alternate alignment where the guanine bulge is positioned at 6’G7’ on the target strand based on structural studies on relabelled 6’U7’ and 6’A7’ bulges on the target strand of Ago ternary complexes reported in this study. The structural studies would predict that the bulged G positioned between 6’ and 7’ would loop out of the duplex and stack on base 1 of the guide strand and that the C6•G6’ pair would adopt a weakened Watson–Crick alignment (labelled by a dashed line), while the A7•C7’ pair would adopt a reversed non-canonical pairing alignment (labelled with an x).
Structure of single base bulge at the cleavage site of the target strand
We next focused our attention from single bulges inserted into the seed segment of the RNA target strand to a single bulge centered about the cleavage site on the RNA target strand.
9’U10’ bulge
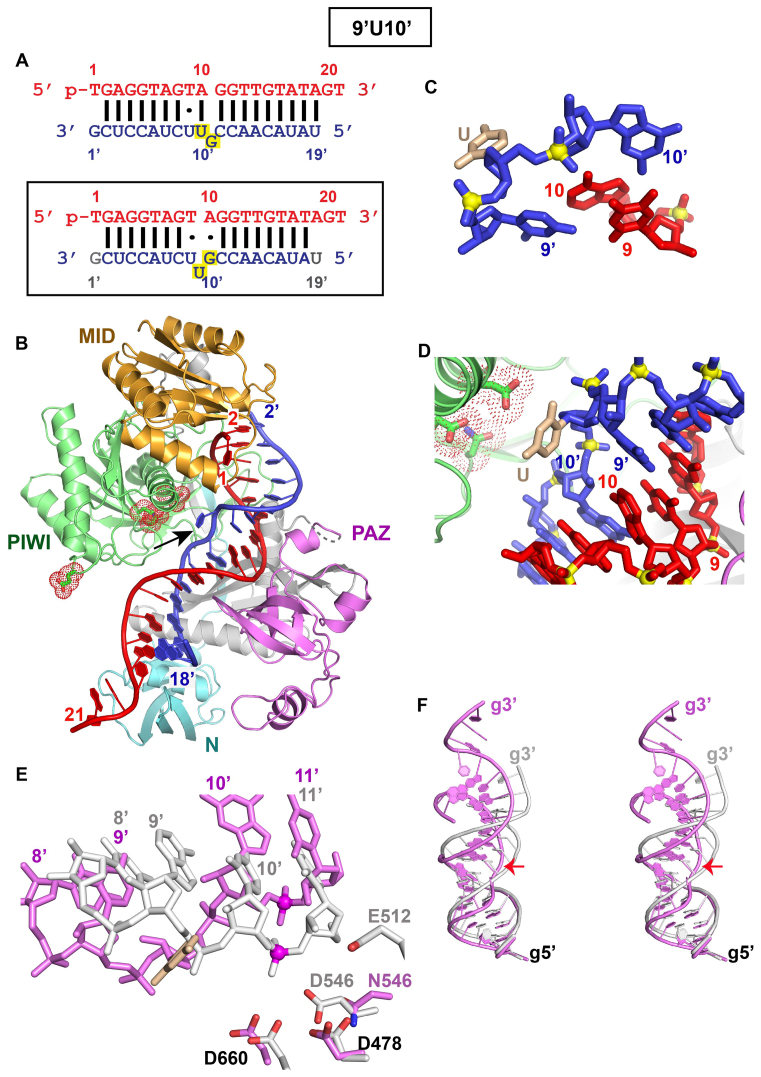
These efforts addressed a TtAgo ternary complex containing a bulged guanine between positions 10’ and 11’ on the target strand (Figure 5A, top panel), but following structural analysis was found instead to contain a bulged uracil between positions 9’ and 10’ on the target strand (Figure 5A, boxed bottom panel). Henceforth, this bulge-containing sequence will be labeled 9’U10’ bulge. We have solved the 2.9 Å crystal structure of the TtAgo ternary complex containing a 9’U10’ bulge (Figure 5B; X-ray statistics listed in Supplementary Table S2), with the 3′-end of the guide strand anchored in the PAZ domain of a second adjacent Ago molecule in the crystal lattice (Supplementary Figure S4A).
Figure 5.
Crystal structure of TtAgo containing D546N catalytic mutant bound to 5′-phosphorylated 21-nt guide DNA and 20-nt target RNA containing a 9’U10’ bulge positioned on the target strand within the cleavage site segment. (A) Sequence of the guide DNA-target RNA duplex (top panel), with the actual alignment of the bulge in the crystal structure of the ternary complex (bottom boxed panel), where an adjacent uracil loops out of the duplex between positions 9’ and 10’ of the target strand. The traceable segments of the nucleotides of the guide DNA and target RNA in the structure of the ternary complex are shown in red and blue, respectively. The dots shown for T9•U9’ and A10•G10’ (lower panel) represent non-canonical pairing alignments, with the latter forming a non-canonical sheared G•A pair. (B) 2.9 Å structure of TtAgo containing N546 catalytic mutant bound to 5′-phosphorylated 21-nt guide DNA (in red) and 20-nt target RNA (in blue) containing a 9’U10’ bulge positioned on the target strand within the seed segment. The black arrow points to the looped out bulge base. There is one molecule of the complex in the asymmetric unit and the 3′-end of the guide strand is inserted into the PAZ pocket of an adjacent molecule in the crystal lattice (not shown). (C) A stick representation of the bulge site spanning T9•U9’ and A10•G10’ non-canonical pairs, with the looped out uracil highlighted in biscuit color in the 9’U10’ bulge-containing ternary complex. (D) A stick representation of the bulge site of the guide-target duplex, with the looped out uracil highlighted in biscuit color in the 9’U10’ bulge-containing ternary complex. Note that the looped out uracil is directed towards the catalytic residues. (E) The positioning of the RNA target strand of the control Ago ternary complex containing no bulge (in silver) and in the 9’U10’ bulge (in magenta) relative to the catalytic residues (D478, D660 and D546N mutant) of the RNase H fold of the PIWI domain in the complex. The catalytic residues are distant from the phosphate linking the 10’-11’ step (colored in magenta) in the 9’U10’ bulge-containing Ago ternary complex (in magenta). (F) Superposition of the guide-target duplex containing no bulge (in silver) and 9’U10’ bulge (in magenta) in Ago ternary complexes. The guide strand is labeled g and the 10’–11’ phosphate at the cleavage site on the target strand is indicated by a red arrow.
The segment spanning the 10’–11’ cleavage site is aligned such that a uracil is looped-out of the helix (Figure 5C, D and stereo view in Supplementary Figure S4B; density is observed for the sugar but not for the base of the looped-out uracil) and is flanked by T9•U9’ and sheared A10•G10’ non-canonical pairs (Supplementary Figure S4C), as shown schematically in Figure 5A, boxed lower panel (dots represent non-canonical pairs). The positioning of the cleavable phosphate (colored in red) in the 9’U10’ bulge Ago ternary complex (in magenta, Figure 5E) is compared with its counterpart in the no-bulge control guide-target duplex (in silver color, Figure 5E). There is considerable distortion of the target strand at and in the vicinity of the 10’–11’ cleavage site, with the cleavable phosphate shifted further away from the catalytic triad in the 9’U10’ bulge-containing ternary complex (in magenta, Figure 5E) compared to its control counterpart ternary complex (in silver, Figure 5E). The extent of conformational perturbation can be compared in a stereo view following superposition of the guide-target duplex in Ago ternary complexes containing the 9’U10’ bulge (in magenta, Figure 5F; g stands for guide strand and red arrow indicates cleavage site) compared with the control guide-target duplex lacking the bulge (in silver, Figure 5F). Such a perturbation of the duplex towards the 3′-end of the guide can also be observed in surface views of Ago in control and 9’U10’ bulge-containing ternary TtAgo complexes of Supplementary Figure S4D and E (see boxed regions), respectively.
Cleavage activity of TtAgo ternary complexes containing bulges within the seed segment of the guide strand
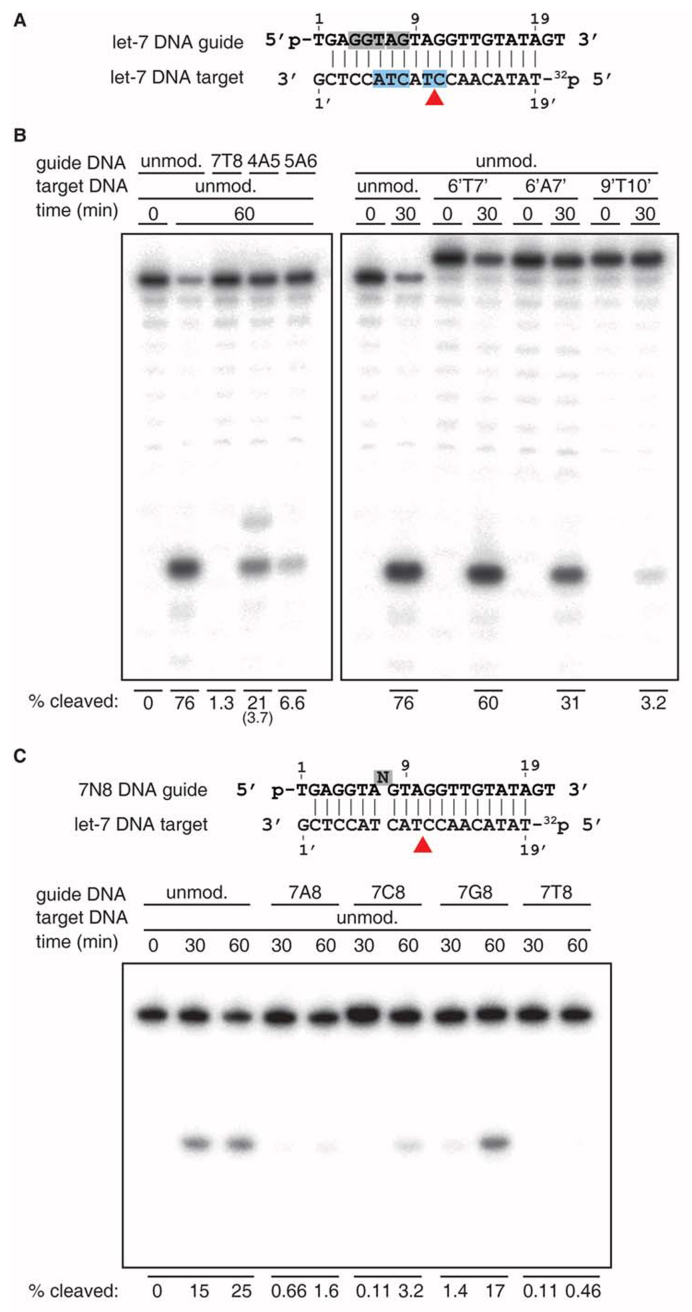
To determine the effect of structurally characterized bulges on cleavage activity, we performed in vitro cleavage assays for TtAgo ternary complexes at 75°C considering that cleavage at 35°C or 55°C was less effective (8,10). We initially studied single-base insertions in the guide strand within the seed segment of a DNA guide-DNA target context as indicated by shaded silver segment in Figure 6A. While insertion of adenosine 4A5 or 5A6 resulted in a 3.6- and 12-fold reduction of cleavage activity at the canonical site between bases 10’ and 11’, respectively, insertion of thymidine 7T8 did not result in detectable cleavage (Figure 6B, left panel). Guide 4A5 also produced a minor cleavage product, one nucleotide longer than the canonical product. While the cleavage activity for 4A5 and 5A6 was predicted from structural results, the absence of cleavage for 7T8 bulged guide was not anticipated. To evaluate the unexpected loss of activity for guide 7T8, for which the structural analysis revealed that the bulged nucleotide could be stacked within the duplex and lead to minimal distortion near the cleavage site, we examined the effect of all other possible nucleotide insertions at this position (Figure 6C). These substitutions showed a surprising range of activity. While guide 7G8 cleavage activity was merely 1.5-fold reduced, guides 7C8 and 7A8 showed a 7.8- and 16-fold reduction, respectively. We also investigated the effect of temperature on cleavage activity by 7T8, hypothesizing that the ternary complex with the bulged guide might have a lower free energy at a lower temperature. However, we did not note any detectable cleavage activity at 55°C. Prolonged incubation for 3 hours at either 55°C or 75°C did not increase the yield of cleaved target (data not shown).
Figure 6.
Effect of bulges on DNA cleavage. (A) Schematic representation of guide and target DNAs. The guide strand 5′ phosphate and the radiolabeled 32P-phosphate of the target strand are indicated. The locations of nucleotide insertions in guide and target strands are in-between gray and blue highlighted nucleotides, respectively. The red arrowhead indicates the canonical cleavage site located between position 10’ and 11’ of the target strand. (B) Effect of bulges on DNA cleavage. TtAgo was pre-incubated with 5′ phosphorylated guide DNAs at 55°C for 30 min prior to addition of 5′ radiolabeled DNA substrate, followed by incubation at 75°C for indicated times. Cleavage products were resolved on a 15% denaturing polyacrylamide gel. The bulge positions are indicated according to structural studies. The fraction of target cleaved was quantified by phosphorimaging and shown at the bottom. The cleavage fraction corresponding to the minor product observed for the 4A5 guide is indicated in parenthesis below the fraction of the major product. (C) Effect of the identity of guide insertions on DNA cleavage. Cleavage assays were performed for every possible insertion between positions 7 and 8 on the guide DNA. The reduction in TtAgo activity in comparison to (C) is due to an additional freeze-thaw cycle of the TtAgo enzyme, which results in partial loss of activity.
As the identity of the inserted nucleotide between guide positions 7 and 8 affected cleavage activity differently (Figure 6C), we also examined the effect of every possible nucleotide insertion at 4N5 and 5N6 sites on the guide strand in a DNA guide-DNA target context (Supplementary Figure S5A and S5B). Bulges 4N5 not only reduce cleavage activity, but also affect accuracy as indicated by the appearance of minor cleavage products indicated by gray arrowheads, regardless of the identity of the inserted nucleotide (Supplementary Figure S5A). Bulge 5N6 substantially reduce cleavage activity and minor cleavage products, if generated, remain undetectable (Supplementary Figure S5B). Our results confirm that there is a nucleotide dependence on the efficiency of cleavage, without any clear a priori preference for specific nucleotides.
Cleavage activity of TtAgo ternary complexes containing bulges within the seed segment and adjacent to the cleavage site of the target strand
We next studied single-base insertions on the target strand within the seed segment and adjacent to the cleavage site in a DNA guide–DNA target context as indicated by shaded cyan segment in Figure 6A. Overall, bulges in the seed region of target DNA preserved cleavage between bases 10’ and 11’ (Figure 6B, right panel). Target 6’T7’ showed a mere 1.3-fold reduction in cleavage efficiency compared to unmodified let-7 target, while target 6’A7’ cleavage was reduced 2.5-fold. Target 9’U10’, with placement of the bulge directly adjacent to the cleavage site, resulted in drastic reduction of activity (24-fold). Although cleavage activity for 6’T7’ and 6’A7’, and lack of cleavage for 9’U10’ were expected from structural observations, the position of the cleavage site for 6’U7’ and 6’A7’ between bases 10’ and 11’ was not.
We also examined the effect of every possible nucleotide insertion at 6’N7’ and 7’N8’ sites on the target strand in a DNA guide-DNA target context (Supplementary Figure S6A and S6B). Our results confirm that there is a nucleotide dependence on the efficiency of cleavage, without any clear a priori preference for specific nucleotides.
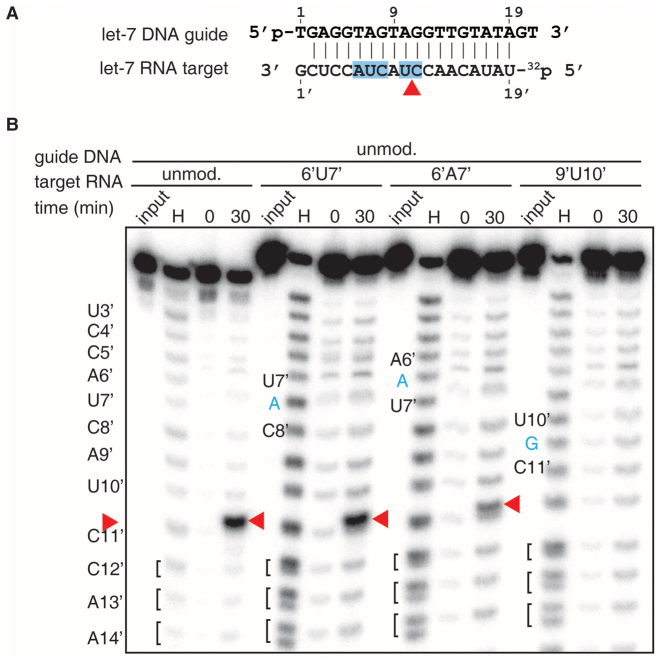
We also performed cleavage assays on RNA targets with bulges within the seed segment or adjacent to the cleavage site in a DNA guide-RNA target context (Figure 7A), as used for structural studies reported in earlier sections. Since RNA is susceptible to divalent-cation-mediated hydrolysis, especially at higher temperatures, we reduced incubation times to minimize hydrolytic degradation. Similar to DNA targets (Figure 6B, right panel), bulges in the seed region of the target RNA (6’U7’ and 6’A7’) directed cleavage between bases 10’ and 11’ (Figure 7B, red arrowheads), and a bulge adjacent to the cleavage site abolished activity.
Figure 7.
Effect of bulges on RNA cleavage. (A) Schematic representation of guide DNA and target RNAs. (B) TtAgo was pre-incubated with 5′ phosphorylated guide DNA at 55°C for 30 min prior to addition of 5′ radiolabeled RNA substrates, followed by incubation at 75°C for indicated times. Products were resolved on a 15% denaturing polyacrylamide gel. Red arrowheads indicate the cleavage site. The sequence of let-7 is shown on the left to annotate the hydrolysis and cleavage positions. The bulge positions are indicated according to structural studies and inserted nucleotides are shown in blue. H, alkaline hydrolysis ladder of 5′ labeled target RNA. Hydrolysis yields terminal 2′,3′-cyclic phosphate, which further hydrolyze to 2′- and 3′-monophosphate. These distinctly charged products resolve towards the bottom of the gel into double bands and are grouped by brackets. TtAgo cleavage products carry a 3′-OH and therefore migrate slower than the RNA hydrolysis products of the same sequence.
DISCUSSION
Though attempts were made to introduce single base bulges at various positions on the guide and target strands spanning both the seed segment and the cleavage site in the context of the TtAgo ternary complex, we were only able to grow diffraction quality crystals and solve the structures for a subset of bulge-containing ternary complexes as outlined above in the results section. Despite the limitations of a finite set of structures, important conclusions can be reached on what contributes to formation of a stable bulge site, as well as the range of bulge-induced long-range helical perturbations that could result in altered positioning of the cleavage site.
Stacked-In versus Looped-Out bulge alignments in TtAgo ternary complexes
Single base bulges can either stack-in or be looped-out of the duplex (26). In the case of TtAgo ternary complexes, intermolecular contacts between the guide-target duplex and the Ago protein could contribute to the alignment of the bulged base. There are extensive intermolecular contacts between the seed nucleotides of the guide strand and amino acids of TtAgo in the ternary complex (8,9), severely limiting the space available for accommodating a looped-out base. Therefore, it was anticipated that single base bulges 7T8 (Figure 1A), 4A5 (Figure 1F) and 5A6 (Figure 2A) on the DNA guide strand would most likely stack into the duplex, as observed from the structures of their ternary TtAgo complexes (Figures 1C, G and 2B, respectively).
By contrast, there are no contacts between the seed nucleotides of the target strand and amino acids of TtAgo in the ternary complex (8,9), and hence there is ample space for a bulged base to loop-out of the duplex. It is therefore not surprising that 6’U7’ (Figure 3A) and 6’A7’ (Figure 4A) bulges on the RNA target strand loop-out of the duplex in their TtAgo ternary complexes (Figures 3C and 4B, respectively).
There are contacts between the 10’ and 11’ step at the cleavage site on the target strand and amino acids of TtAgo in the structure of ternary complex (8,9). Hence, if the uracil were to loop out in the 9’U10’ duplex, there would have to be conformational changes to overcome steric clashes with the amino acids of the Ago protein. Indeed, looping out of the uracil within the 9’U10’ bulge resulted in shifting of the 10’–11’ phosphodiester linkage away from the catalytic triad in this bulge-containing duplex (in magenta, Figure 5E) compared to its control counterpart lacking the bulge residue (in silver, Figure 5E).
Structure-based bulge-induced long-range helical perturbations and altered positioning of cleavage site
We observe a range of bulge-induced long-range helical perturbations and altered positioning of the cleavage site in the structures of the bulge-containing TtAgo ternary complexes reported in this study. Unexpectedly, the seed segment 7T8 bulge on the guide strand exhibited the least perturbation (Figure 1E; 7T8 in blue and control in silver) and the relative positions of the cleavable 10’–11’ phosphate on the target strand and the catalytic tetrad were essentially indistinguishable from their control counterpart lacking the bulge residue (Figure 1D; 7T8 in blue and control in silver). The T residue of 7T8 of the guide strand stacked into the duplex and the conformational adjustments appear to be localized and did not extend to the cleavage site. The same stacked-in alignment and minimal conformational perturbation at the cleavage site was also observed for 4A5 (Figure 1G, H) and 5A6 (Figure 2B, C) bulges on the DNA guide strand in TtAgo ternary complexes. Indeed, cleavage was observed at the 10’–11’ step of the target strand in the crystal structure of the 4A5 bulge-containing DNA guide–DNA target duplex in the presence of wild-type TtAgo (Figure 1H and Supplementary Figure S2D).
By contrast, a pronounced conformational change was observed at the cleavage site for the 6’U7’ and 6’A7’ bulges within the seed segment on the RNA target strand, where the bases are looped-out of the helix. In both these cases, a notable widening of the groove and a pronounced conformational change in the PAZ domain was observed relative to the control duplex (shown for 6’U7’ bulge in Figure 3I). This transition was accompanied in both cases by repositioning of the target strand sugar-phosphate backbone, such that the 11’–12’ step rather than the 10’–11’ step, is positioned opposite the catalytic tetrad in the 6’U7’ (Figure 3H) and 6’A7’ (Figure 4F) bulge TtAgo ternary complexes.
The largest conformational change was observed for the 9’U10’ bulge (Figure 5A, bottom panel) flanking the cleavage site on the target strand. The U is looped out of the helix and directed towards the catalytic residues, such that the resulting perturbations contribute to an increased separation of the backbone phosphate linking positions 10’ and 11’ on the target strand from the catalytic triad residues (Figure 5E). Indeed, the trajectory of the guide DNA–target RNA duplex changes dramatically on proceeding from the control duplex (Figure 5F, in silver) to the 9’U10’-bulge containing duplex (Figure 5F, in magenta).
Cleavage-compatible conformations adopted by seed segment bulge-containing TtAgo ternary complexes
Our previous structural studies on TtAgo complexes with bound 5′-phosphoryated DNA guide strand to form binary complexes (7) and with added RNA target strands of variable length to form ternary complexes (8) (highlighted the need to release the 3′-end of the guide from the PAZ pocket with increased RNA target length so as to switch TtAgo from a cleavage-incompatible to a cleavage-compatible conformation. The same was later shown to hold TtAgo ternary complexes containing guide DNA and target DNA strands (9). This criterion cannot be used in the current bulge-containing TtAgo ternary complexes since the 5′-phosphate and 3′-ends of the guide strand are not anchored in MID and PAZ pockets respectively of the same Ago protein for the 6’U7’ (Supplementary Figure S3A), 6’A7’ and 9’U10’ (Supplementary Figure S4A) bulges on the target strand.
Recently, it has been shown that a Glu adopts an external ‘cleavage-incompatible’ conformation but inserts into the catalytic pocket to complete a catalytic tetrad on generation of a ‘cleavage-compatible’ conformation (9,12). We observe the catalytic Glu inserted into the catalytic pocket in the 6’U7’ (Figure 3B) and 6’A7’ (Supplementary Figure S3F), as well as in the 7T8 (Figure 1B), 4A5 (Supplementary Figure S2A), and 5A6 (Supplementary Figure S2E) bulge-containing ternary complexes indicative of generation of cleavage-compatible conformations. By contrast, in the 9’U10’ bulge-containing Ago ternary complex, the Glu is directed outwards from the catalytic pocket (Figure 5B), indicative of a ‘cleavage-incompatible’ conformation.
Factors contributing to stable conformation for seed segment bulges on guide strand
Our structural studies have established that 7T8 (Figure 1C), 4A5 (Figure 1G) and 5A6 (Figure 2B) bulges positioned within the seed segment of the guide strand all stack into the duplex with minimal perturbation in TtAgo ternary complexes. Such alignments are stabilized by stacking of the inserted bulged base with somewhat weakened adjacent Watson–Crick base pairs. Such a stacked-in alignment involves localized conformational changes and minimally perturbs the intermolecular contacts between the seed segment of the guide strand and the backbone and side chains of the Ago scaffold.
Factors contributing to stable conformation for 6’N7’ bulge on target strand
Our structural studies have identified a stable conformation adopted by bulge bases at 6’U7’ (Figure 3A, bottom panel) and 6’A7’ (Figure 4A, bottom panel) steps spanning the seed segment of the target strand in TtAgo ternary complexes. The common theme in these structures includes looping-out of the base between positions 6’ and 7’ of the target strand in both cases (Figures 3D, E and 4C, D), in the process retaining the weakened Watson–Crick base pair in the flanking 6–6’ position, but generating a reversed non-canonical alignment in the flanking 7•7’ position (Figures 3A and 4A, boxed bottom panels). Such adjacent positioning of locally anti-parallel (Watson–Crick pair) and parallel (reversed non-canonical pair) helical segments facilitates looping out of the base positioned between them (Figures 3C and 4B).
The 6’U7’ (Figure 3C–F) and 6’A7’ (4B–E) bulges positioned within the seed segment of the target strand, adopt very similar conformations in their TtAgo ternary complexes. The single base bulges are looped out of the target strand and stack on the unpaired first base of the guide strand, with both stacking and hydrogen bonding stabilizing this alignment (Figures 3E and 4D). These structural contributions on 6’N7’ bulges in eubacterial TtAgo ternary complexes can be compared with proposed stable G-bulge sites within the seed segment of target strands identified from a genome-wide map of miRNA interaction sites in mouse brain using the HITS-CLIP approach (33). Essentially, these authors noted the high propensity of G-bulge sites spanning position 5’G6’ on the target strand as shown for the 5′-UGGCCUU-3′ motif matched to miRNA-124 (Figure 4G). However, our structural studies imply consideration of an alternate alignment, whereby the G-bulge site could span position 6’G7’ (Figure 4H, boxed bottom). Our structural studies outlined in this paper on 6’U7’ and 6’A7’ bulges in the context of the TtAgo ternary complexes, favor the 6’G7’ alignment over the earlier proposed 5’G6’ alignment (33). In addition, based on our structural studies, we would predict that the G-bulge would loop out (between positions 6’ and 7’) of the duplex and be flanked by a weakened Watson–Crick C6–G6’ pair (Figure 4H; dashed line represents a weakened pair) and a reversed A7•U7’ pair (Figure 4H, x symbol represents reversed alignment). It should be pointed out that our structural studies on eubacterial TtAgo ternary complexes involve guide DNA and target RNA, while the earlier HITS-CLIP studies on eukaryotic mouse Ago ternary complexes involved guide RNA and target RNA (33).
Cleavage activity and position in bulge-containing TtAgo ternary complexes
Prior structural and functional studies established that TtAgo uses a 5′ phosphorylated guide DNA to cleave both DNA and RNA targets (8–10). However, the accommodation of bulges as they may arise in hybridization to imperfectly complementary targets has not been studied in this particular system readily amenable to structural and functional studies. In mammalian Ago systems a G bulge at position 6’ was well tolerated for specific miRNA-target RNA seed interactions (33), yet in vivo biological effects of such non-canonical pairing in the broader context of many miRNAs appear much less or not at all important (34,35). These observations prompted us to study the accommodation of nucleotide bulges by TtAgo and the downstream effects on cleavage activity and cleavage position.
Cleavage assays revealed that irrespective of the sequence and position of the bulge a reduction in cleavage activity was observed. However, the loss in activity varied widely from a mere 1.3-fold reduction to undetectable cleavage, equivalent to a greater than 50-fold reduction, as defined by the detection range of our assay. Our functional analysis focused on bulge positions with structural information available, including nucleotide positions 4–6 and 7–8 for the guide, and 6–8 and 9–10 for the target. Among the bulge-accommodating structures predicting activity by not perturbing the geometry of the cleavage site, several interesting exceptions were encountered that argue for a complex interplay of kinetic and thermodynamic factors ultimately responsible for the occasional discrepancies between structural and biochemical studies.
The 7T8 guide bulge did not support cleavage (Figure 6), while the structure revealed that it was accommodated within the stacked helix without propagating distortions affecting the geometry of the cleavage site or cleavage position. However, when we tested the influence on activity of other nucleotides at this position, we were surprised to obtain a nearly complete rescue of activity for bulge 7G8, followed by a 7.8-fold reduction for 7C8, while activity for 7A8 was even further reduced (Figure 6C). This indicated that there was no clear preference for accommodating purines over pyrimidines at this position, even though purines may offer a larger surface for duplex-stabilizing stacking interactions. It is important to also keep in mind that the temperatures at which crystals are formed are well below the temperature we can detect cleavage activity.
The accommodation of 6’A7’ and 6’U7’ target strand bulges in our structures predicted cleavage, with positioning of the scissile phosphate one nucleotide downstream of the canonical cleavage site when counting from the 5′ end of the guide. While cleavage was indeed only slightly reduced for both bulged targets, the cleavage site remained the same as for unmodified target. We currently have no explanation for this discrepancy related to cleavage position on the target strand between conclusions reached from structural and enzymatic studies.
In only one instance, with guide bulge 4A5 and unmodified target, did we observe the simultaneous appearance of a canonical and a minor alternative cleavage site (Figure 6B, left panel), supporting the general structural finding that accommodation of bulges can also lead to repositioning of the cleavage site. Surprisingly, the structure in this case (Figure 1G) showed no indication for alternative positioning of the cleavage site. Together, these findings indicate that the cleavage site position for bulged duplexes cannot always be readily inferred from structural studies and requires biochemical analysis. They should also caution computational biologists for adapting rules predicting biological activity for bulged miRNA–mRNA duplexes involving Ago proteins.
In summary, our biochemical studies expand the guide and target repertoire supporting Ago-protein-mediated cleavage using bulged guides and targets. We observed a wide range of sequence- and position-dependent activity not always in complete agreement with structural studies, thus underscoring the importance of functional assays.
ACCESSION NUMBERS
The structures of ternary complexes of T. thermophilus Ago have been deposited to the Protein Data Bank. The accession codes are: Ternary TtAgo DNA guide-DNA target complexes containing 7T8 (5XOU), 5A6 (5XQ2) and 4A5 (5XP8) bulges on the guide strand. Ternary TtAgo DNA guide-RNA target complexes containing 6’U7’ (5XPG), 6’A7’ (5XOW) and 9’U10’ (5XPA) bulges on the target strand. All TtAgo ternary bulge-containing complexes contained a catalytically inactive D546N mutant, except for the 4A5 bulge, where the TtAgo protein was wild-type protein.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the staff of beamline BL-17U at Shanghai Synchrotron Radiation Facility (SSRF), beamlines BL-1A and BL-17A at Photon Factory and NE-CAT beam lines at the Advanced Photon Source (APS), Argonne National Laboratory, and the X-29 beamline at the Brookhaven National Laboratory, supported by the US Department of Energy, for assistance with data collection.
Author contributions: G.S. and Y.W. expressed and purified wild-type T. thermophilus Ago and its catalytic mutants, as well as grew crystals of the various ternary complexes. Y.W. solved the structures of these ternary complexes. T.G., A.S. and S.J. undertook the cleavage assays under the supervision of T.T., D.P., T.T. and Y.W. supervised the project and wrote the paper with all authors having read and approved the submitted manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [31571335, 31630015, 91440201]; Chinese Ministry of Science and Technology [2014CB910102 to Y.W.]; NIH [TR01 GM104962-01 to D.J.P. and T.T.]. Funding for open access charge: Natural Science Foundation of China [31571335].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dykxhoorn D.M., Novina C.D., Sharp P.A.. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003; 4:457–467. [DOI] [PubMed] [Google Scholar]
- 2. Baulcombe D. RNA silencing in plants. Nature. 2004; 431:356–363. [DOI] [PubMed] [Google Scholar]
- 3. Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005; 122:17–20. [DOI] [PubMed] [Google Scholar]
- 4. Rana T.M. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007; 8:23–36. [DOI] [PubMed] [Google Scholar]
- 5. Hutvagner G., Simard M.J.. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008; 9:22–32. [DOI] [PubMed] [Google Scholar]
- 6. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J.. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008; 456:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J.. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J. et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olovnikov I., Chan K., Sachidanandam R., Newman D.K., Aravin A.A.. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. cell. 2013; 51:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J.. Structure of yeast Argonaute with guide RNA. Nature. 2012; 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakanishi K., Ascano M., Gogakos T., Ishibe-Murakami S., Serganov A.A., Briskin D., Morozov P., Tuschl T., Patel D.J.. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 2013; 3:1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J.. Structural basis for 5΄-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005; 434:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parker J.S., Roe S.M., Barford D.. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005; 434:663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma J.B., Ye K., Patel D.J.. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004; 429:318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lingel A., Simon B., Izaurralde E., Sattler M.. Nucleic acid 3΄-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004; 11:576–577. [DOI] [PubMed] [Google Scholar]
- 20. Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L.. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004; 305:1434–1437. [DOI] [PubMed] [Google Scholar]
- 21. Parker J.S., Roe S.M., Barford D.. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004; 23:4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J.. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. cell. 2005; 19:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuhn C.D., Joshua-Tor L.. Eukaryotic Argonautes come into focus. Trends Biochem. Sci. 2013; 38:263–271. [DOI] [PubMed] [Google Scholar]
- 24. Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V., Patel D.J., van der Oost J.. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014; 21:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ipsaro J.J., Joshua-Tor L.. From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 2015; 22:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hermann T., Patel D.J.. RNA bulges as architectural and recognition motifs. Structure. 2000; 8:R47–R54. [DOI] [PubMed] [Google Scholar]
- 27. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 28. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 29. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 31. Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 32. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 33. Chi S.W., Hannon G.J., Darnell R.B.. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 2012; 19:321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Friedman R.C., Farh K.K., Burge C.B., Bartel D.P.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nam J.W., Rissland O.S., Koppstein D., Abreu-Goodger C., Jan C.H., Agarwal V., Yildirim M.A., Rodriguez A., Bartel D.P.. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. cell. 2014; 53:1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.