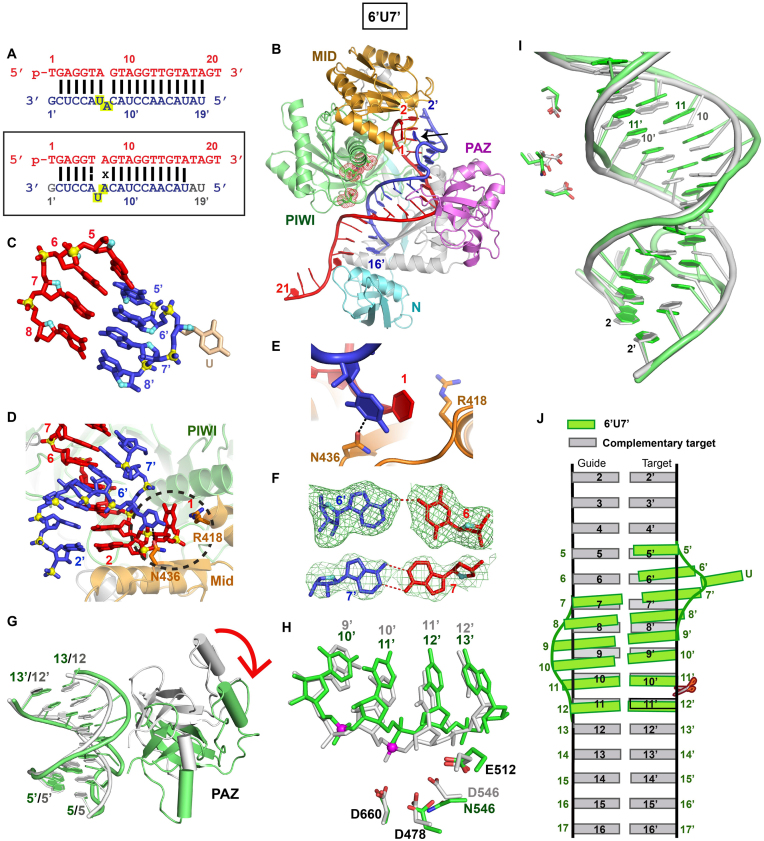
Figure 3.
Crystal structure of TtAgo (D546N catalytic mutant) bound to 5′-phosphorylated 21-nt guide DNA and 20-nt target RNA containing a 6’U7’ bulge positioned on the target strand within the seed segment and conformational adjustments on proceeding from control to 6’U7’ bulge-containing ternary complexes. (A) Sequence of the intended guide DNA–target RNA duplex (top panel), with the actual alignment of the bulge in the crystal structure of the ternary complex (bottom boxed panel), where an adjacent uracil loops out of the duplex between positions 6’ and 7’ of the target strand. The traceable segments of the nucleotides of the guide DNA and target RNA in the structure of the ternary complex are shown in red and blue, respectively. The dashed line for the T6–A6’ pair represents a weakened Watson–Crick pair stabilized by one hydrogen bond, while x shown for the A7•A7’ pair represents a reversed non-canonical pairing alignment. (B) 2.8 Å structure of TtAgo (D546N catalytic mutant) bound to 5′-phosphorylated 21-nt guide DNA (in red) and 20-nt target RNA (in blue) containing a 6’U7’ bulge positioned on the target strand within the seed segment. The black arrow points to the looped out bulge base. There is one molecule of the complex in the asymmetric unit and the 3′-end of the guide strand is inserted into the PAZ pocket of an adjacent molecule in the crystal lattice (see Supplementary Figure S3A). (C) A stick representation of the bulge site and two flanking base pairs on either side, with the looped out uracil highlighted in biscuit color in the 6’U7’ bulge-containing ternary complex. Note the opposing directionalities of the sugar ring oxygens (in cyan) on either side of the bulge site. (D) An overall view highlighting the looped-out uracil of the target RNA and its stacking on sheared-apart base 1 of the guide DNA in the 6’U7’ bulge-containing ternary complex. (E) Stacking of the looped our uracil with unpaired first base of the guide strand and hydrogen bonding with the side chain of Asn436 in the 6’U7’ bulge-containing ternary complex. (F) An omit map (3σ) identifying pairing alignment for the weakened Watson–Crick A6•U6’ pair (top panel) and the reversed A7•A7’ pair (bottom panel) flanking the bulged looped out uracil in the 6’U7’ bulge-containing ternary complex. (G) Superposition of the PAZ domain and guide-target duplex containing no bulge (in silver) and 6’U7’ bulge (in green) in Ago ternary complexes. A red arrow indicates the large conformational transition observed for the PAZ domain on proceeding from the no bulge (in silver) to 6’U7’ bulge (in green) ternary complexes. (H) The positioning of the RNA target strand in the no bulge-containing control (in silver) and in the 6’U7’ bulge-containing (in green) Ago ternay complexes relative to the catalytic residues of the RNase H fold of the PIWI domain in the complex. The catalytic residues are distant from the phosphate linking the 10’–11’ step (colored in magenta) and closer to the phosphate linking the 11’–12’ step in the 6’U7’ bulge-containing Ago ternary complex (in green). (I) Superposition of the seed and cleavage site segments of the guide-target duplex in the no-bulge control (in silver) and 6’U7’ bulge-containing (in green) Ago ternary complexes. (J) Schematic emphasizing base displacement of the target strand between 5–5′ and 11–11’ pairs in the duplex of the 6’U7’ bulge-containing Ago ternary complex (in green), relative to the duplex of the no-bulge control ternary complex (in silver).