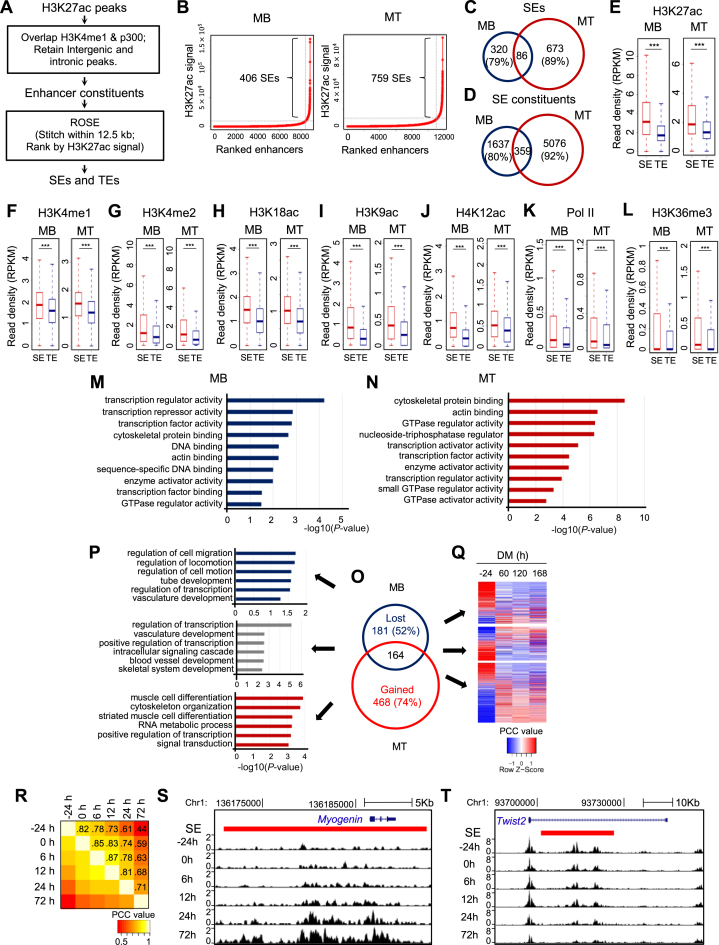
Figure 1.
Identification and characterization of SEs in skeletal muscle differentiation. (A) Illustration of the computational pipeline to identify TEs and SEs in MB and MT. (B) SEs were identified in a stage-specific manner in MB or MT utilizing ROSE, a program that stitches and ranks enhancers. When the background subtracted H3K27ac signal for enhancers was plotted in a ranked order, a clear geometric inflection point was revealed on the curve, with the ones above this point being super-enhancers (SEs), and the rest typical enhancers (TEs). (C and D) Shifted landscapes of SEs and SE constituents from MB to MT. A high portion was lost or gained during the differentiation. (E–L) Increased ChIP-seq intensity of enhancer marks (e.g. H3K27ac, H3K4me1, H3K4me2, H3K18ac, H3K9ac and H4K12ac), and indicators of active transcription (e.g. Pol II and H3K36me3) was observed on SE than TE constituents. ***P < 0.001. (M and N) Gene Ontology (GO) analysis of molecular functions for SE associated genes revealed enrichment of transcription regulator/factor activity in both MB and MT. The y-axis shows the top 10 enriched GO terms and the x-axis shows the enrichment significance P-values. (O) The number of SEs lost, maintained or gained as cells progressed from MB to MT. (P) GO analysis of the genes (top five) associated with the above categories of SEs. (Q) The differential expression patterns of the above genes were examined by RNA-seq collected from MB or MT at various time points in differentiation medium (DM). (R) H3K27ac ChIP-seq signals revealed SE landscape shifts with the progression of differentiation program from −24, 0, 6, 12, 24 and 72 hr in DM. The Pearson correlation coefficient (PCC) values are labeled on each grid to indicate the degree of correlation between the two consecutive stages. (S) Increasing H3K27ac density was observed for the SE (red bar) associated with Myogenin gene during the differentiation. (T) Decreasing H3K27ac density was observed for the SE associated with Twist2 gene during the differentiation.