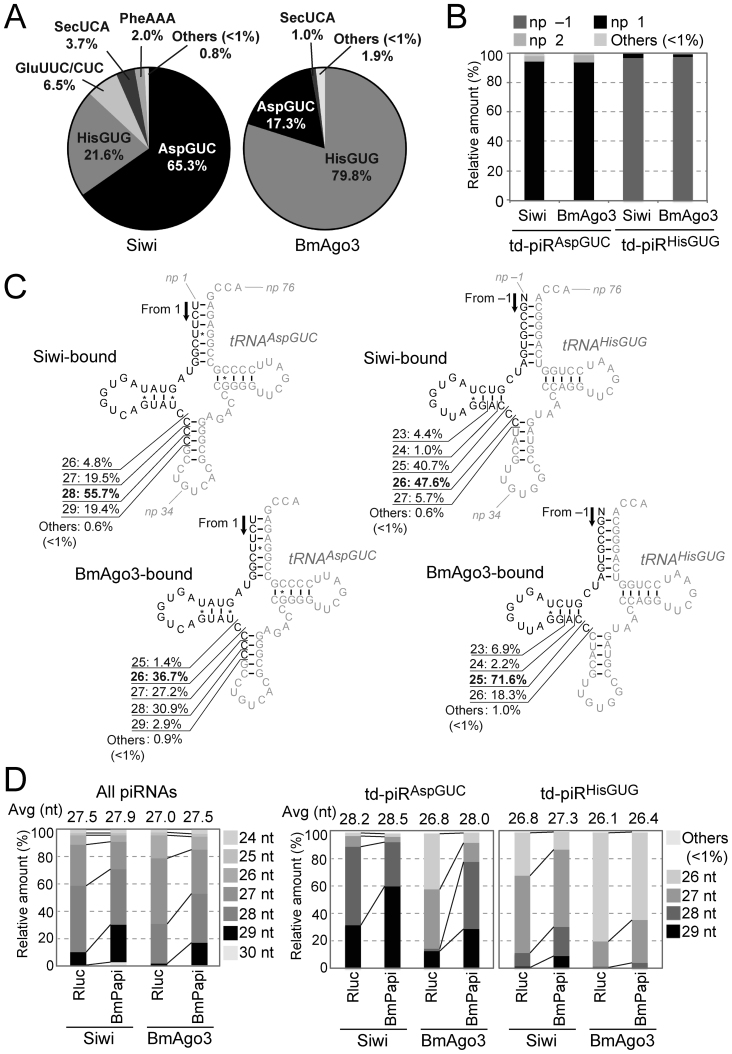
Figure 1.
Identification of td-piRNAs in BmN4 cells. (A) BmN4 Siwi- or BmAgo3-bound piRNAs were mapped against Bombyx mori tRNA sequences. Among the tRNA-mapped piRNA reads, pie charts show the percentages of the reads derived from respective cyto tRNA species. (B) The 5′-terminal position of Siwi- or BmAgo3-bound td-piRAspGUC or td-piRHisGUG in the respective mature tRNA. Nucleotide positions (np) are indicated according to the nucleotide numbering system of tRNAs (2). (C) The regions from which td-piRAspGUC, starting from np 1, or td-piRHisGUG, starting from np –1, were derived are shown in black in the cloverleaf secondary structure of Bombyx cyto tRNAAspGUC-V1 (Supplementary Figure S2) and tRNAHisGUG. Non-piRNA-derived regions are shown in gray. (D) Read-length distribution of the Siwi- or BmAgo3-bound total piRNAs, td-piRAspGUC, or td-piRHisGUG identified in Rluc-depleted (control) or BmPapi-depleted BmN4 cells.