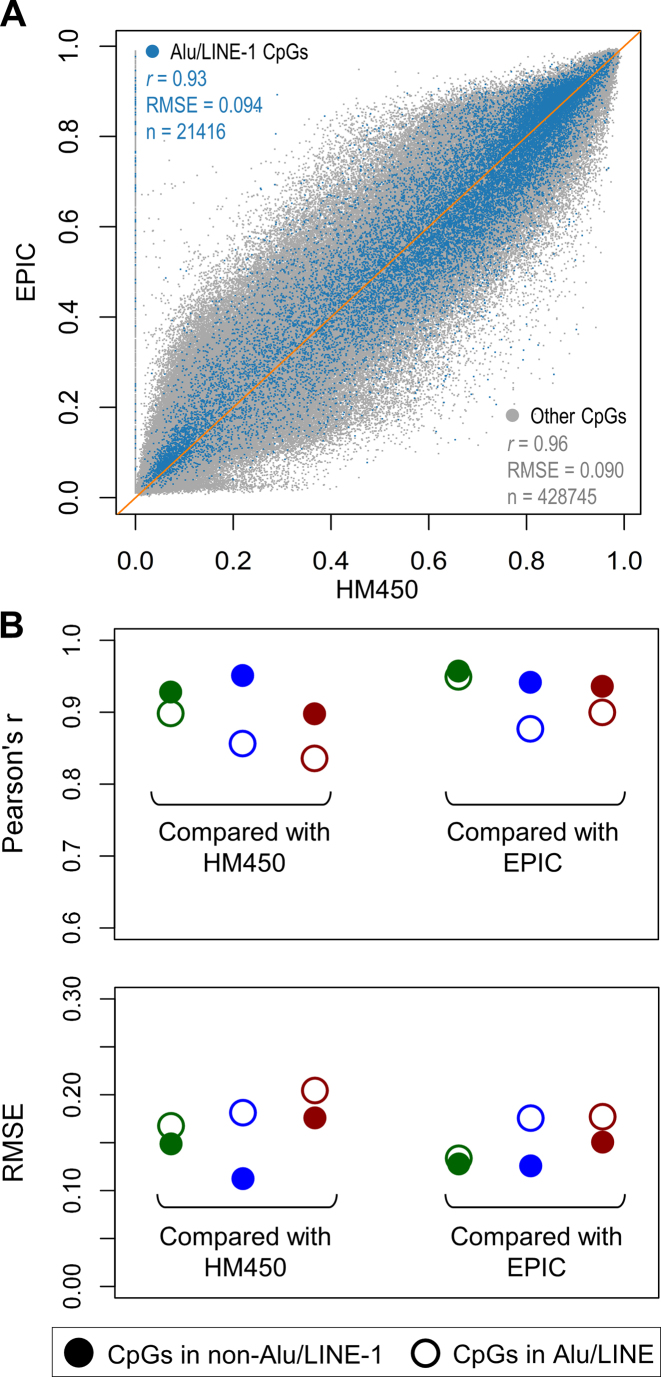
Figure 2.
Reliability of the profiling platforms interrogating CpG sites in Alu and LINE-1. If probes or reads targeting RE regions such as Alu and LINE-1 are affected by ambiguous mapping, methylation readings on these CpGs are more likely to yield different values for the same sample across different platforms. (A) Plot showing high correlation between CpGs profiled using both HM450 and EPIC, with CpGs in Alu/LINE-1 showing slightly smaller r and larger RMSE (root mean square error). (B) Evaluation of the reliability of the three sequencing-based platforms (using Infinium methylation arrays as the benchmark): NimbleGen (green), RRBS (blue), and WGBS (red). NimbleGen shows the highest concordance between both Alu/LINE-1 and non-Alu/LINE-1 CpGs.