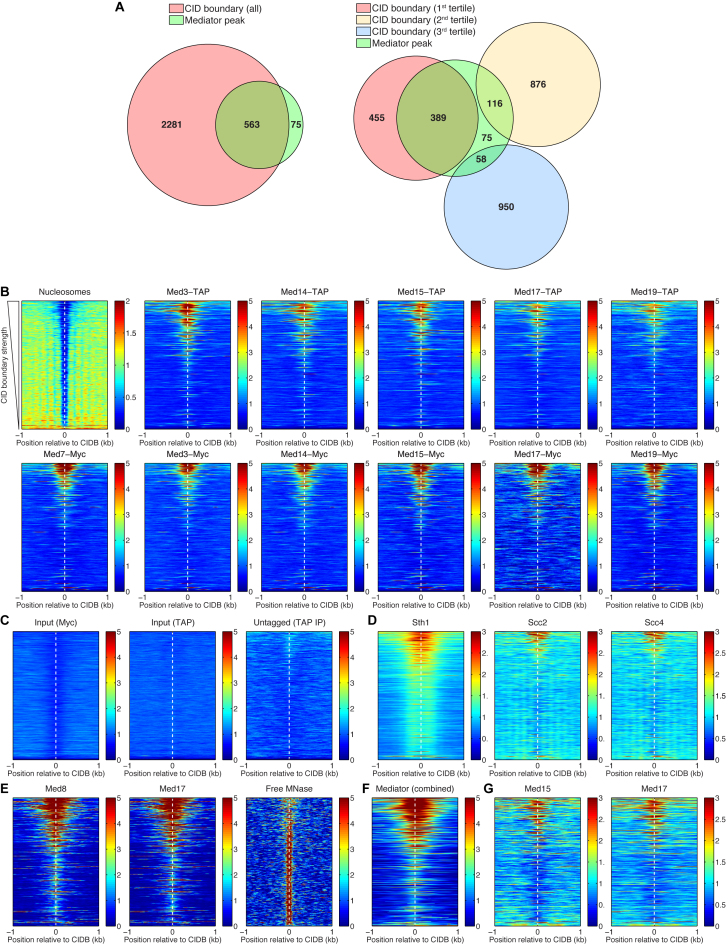
Figure 4.
Mediator, chromatin remodeler RSC, and cohesin loading factors Scc2/Scc4 preferentially bind to the strongest CID boundaries. (A) Venn diagrams representing the overlaps of the CID boundaries and the Mediator binding sites. The CID boundaries are shown as a whole group and split into three tertiles, according to their strength. From the total of 563 Mediator peaks that are found at CID boundaries, about 69% of these are located at the first tertile, containing the strongest CID boundaries. (B) Heat maps representing the nucleosome depletion at all CID boundaries (CIDBs), and the preferential binding of Mediator to the strongest boundaries. The rows of the heat maps are sorted according to strength of the CID boundaries, from the strongest (at the top) to the weakest (at the bottom). (C) Control experiments—sonicated chromatin (input) and IP from an untagged strain—resulted in a clean background, without spurious binding peaks. (D) Sth1 (subunit of the RSC chromatin remodeling complex) and Scc2, Scc4 (cohesin loading factors) also preferentially bind to the strongest CID boundaries. (E) ChEC-seq data for Med8 and Med17 subunits and the control experiment (free MNase cleavage) (59). (F) Combined Mediator ChIP-chip data—MetaMediator Data Set in WT Cells from (58). (G) ChIP-seq data for Med15 and Med17 (55).