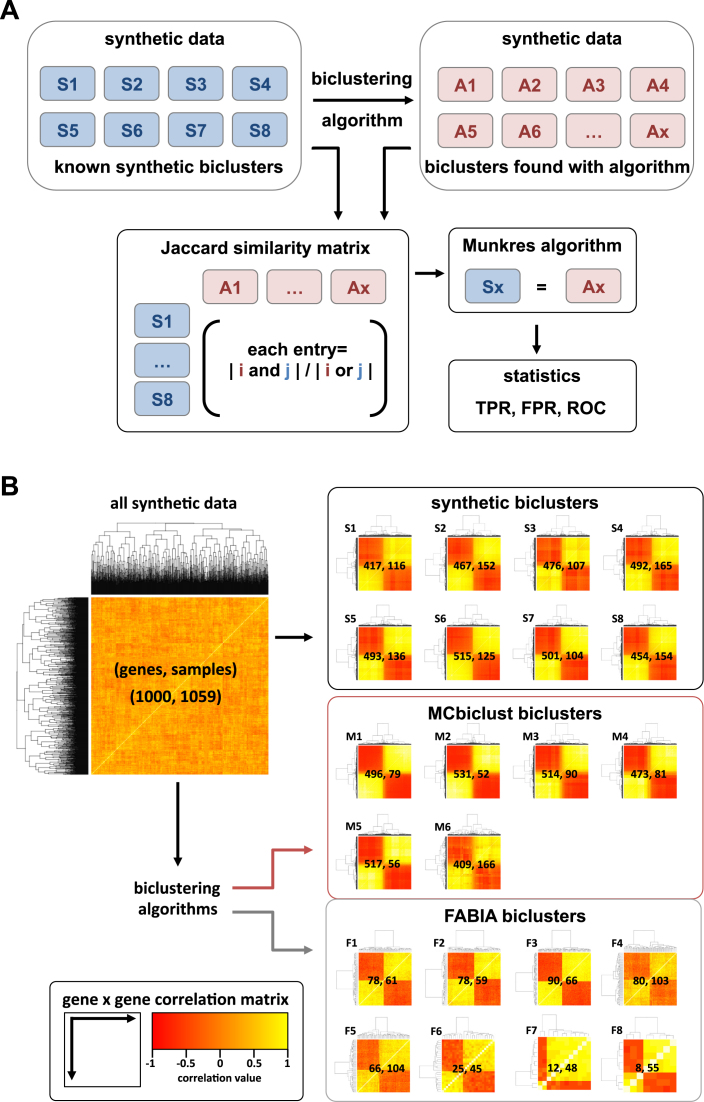
Figure 2.
Benchmarking of MCbiclust against previous biclustering methods. (A) Outline of the evaluation pipeline. Known biclusters in the synthetic datasets are compared with the biclusters found with different biclustering methods. Jaccard Index and the Munkres algorithm is used to solve the assignment problem of matching the known synthetic biclusters with the found biclusters, from which statistical evaluations such as true and false positive rates (TPR, FPR) and relative operating characteristics (ROC) curves are produced. (B) Heatmaps of the gene-gene correlation matrices for all the synthetic data, the known synthetic biclusters (S1–S8) and the biclusters found with FABIA (F1–F8) and MCbiclust (M1–M6). Numbers of gene and samples are shown in parenthesis (gene, sample) to compare the sizes of real biclusters with the ones found with either method.