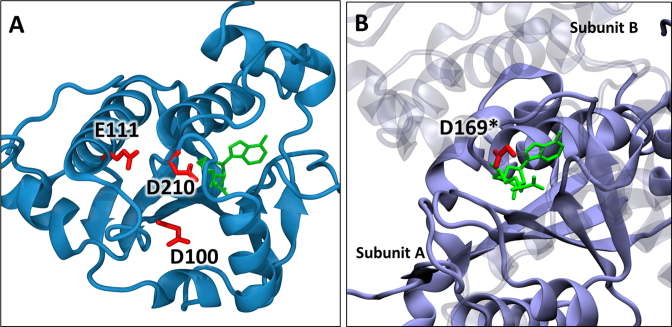
Figure 8.
Locations of conserved carboxylate residues in SPOUT methyltransferases. (A) Crystal structure of Saccharomyces cerevisiae Trm10 (PDB:4JWJ) (25) showing the positions of D210, D100 and E111. The S-adenosyl homocysteine is shown in green. (B) Crystal structure of Haemophilus influenzae TrmD (PDB: 4YVI) (40) showing the active site composed of residues from two subunits. The SAM analog (sinefungin in green) is bound to subunit A (opaque) and the catalytic residue D169* of subunit B (transparent) is shown in red.