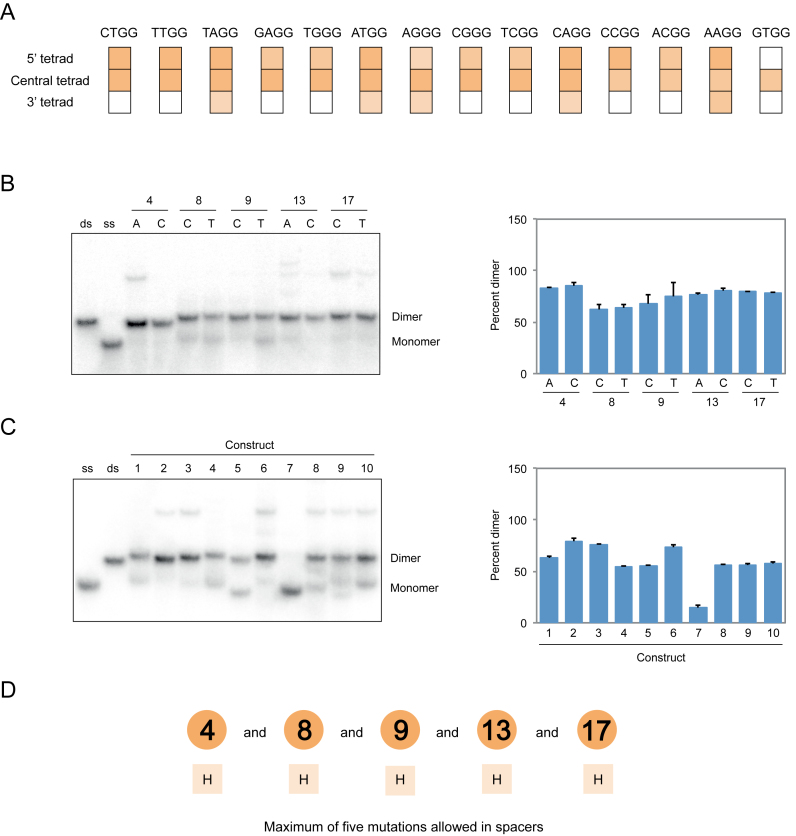
Figure 6.
Sequence model for dimeric G-quadruplexes. (A) Effect of transplanting mutations that induce dimer formation when present in the central tetrad of the reference construct to the 5′ tetrad (nucleotides 1, 5, 10 and 14) or the 3′ tetrad (nucleotides 3, 7, 12 and 16). Colors match those in Figure 4. (B) Native gel and graph showing effects of point mutations in loops on dimer formation. (C) Native gel and graph showing the ability of ten randomly chosen sequences with up to five mutations in loops to form dimers. Sequences are given in Supplementary Table S1. (D) Loop requirements of G-quadruplex variants that form dimers. Numbers indicate the positions of loops in the reference construct, and letters below indicate the nucleotides that can occur at each position. Experiments were performed at 10 μM DNA concentration in a buffer containing 200 mM KCl, 1 mM MgCl2 and 20 mM HEPES pH 7.1. H = A, C or T. Mutations in panels B and C were made in the context of a construct with an AGGG mutation in the central tetrad of the reference construct with the sequence GAGTGGGAAGGGTGGGA.