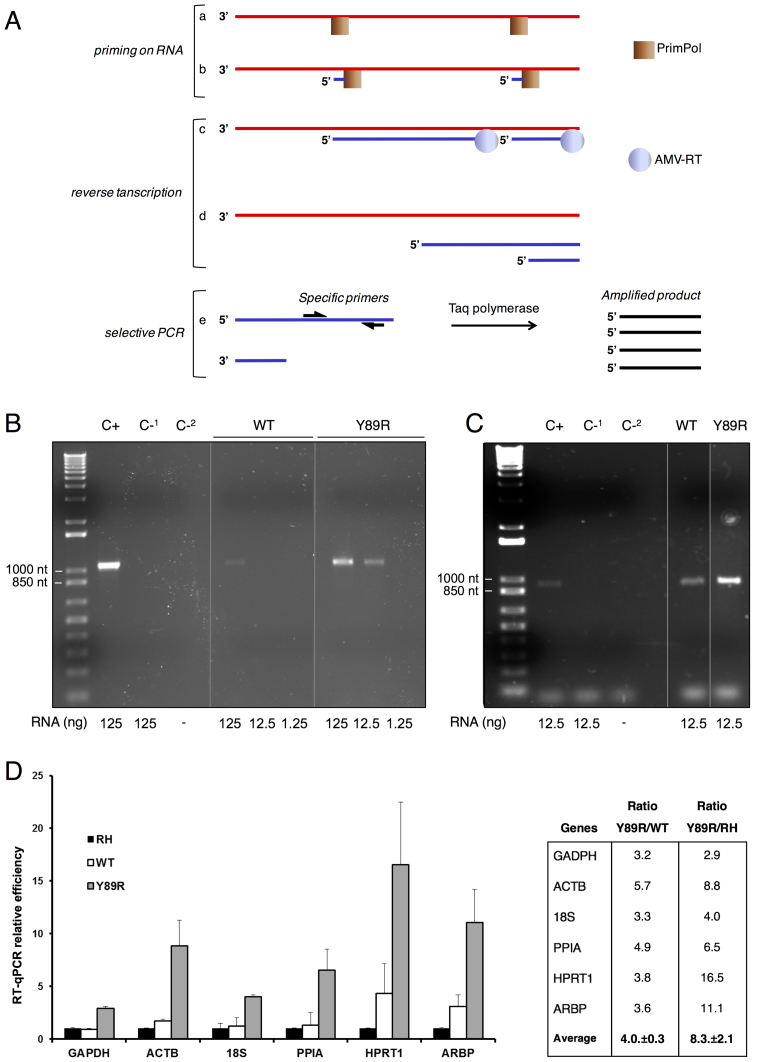
Figure 6.
RT-PCR of RNA primed by HsPrimPol. (A) Scheme of the RT-PCR reaction carried out with HsPrimPol. Red lines are RNA, blue lines are ssDNA, black lines are dsDNA. a: binding of HsPrimPol to the RNA, b: synthesis of DNA primers. c & d: synthesis of ssDNA by AMV-RT. e: PCR reaction with specific primers and synthesis of the dsDNA amplicon. (B) RT-PCR amplification of NS5A HCV gene using WT HsPrimPol or Y89R variant to produce primers during the first reaction step. The amount of RNA template is indicated above. Positive RT-PCR control (C+) using the corresponding reverse primer during the RT process, and negative RT-PCR controls in the presence. (C-1) or absence (C-2) of RNA, are indicated. Molecular size markers and the corresponding sizes (base pairs) are indicated on the left. Size of the expected amplicon is 1051 nt. (C) RT-PCR amplifications of NS3 HCV gene using HsPrimPol or Y89R HsPrimPol to produce primers during the first reaction step. Amount of RNA templates is indicated. Positive RT-PCR control (C+) using random decamers to prime the RT, and negative controls (C-1 and ‘C-2, as in B), are indicated. Size of the expected amplicon is 937 nt. (D) Y89R mutant renders higher RT-qPCR efficiency. The efficiency of DNA synthesis in RT-qPCR reactions primed with either WT (white), or Y89R HsPrimPol (gray), for six different housekeeping genes (see Materials and Methods), is calculated from the Cq values and compared to RT-qPCR primed with random hexamers normalized to 1 (‘RH’; black). For the indicated genes, the ratio of DNA produced in the RT-qPCR primed with Y89R variant respect to that primed with WT HsPrimPol or RH is shown. Averages for each set of ratios, with the standard errors, are shown.