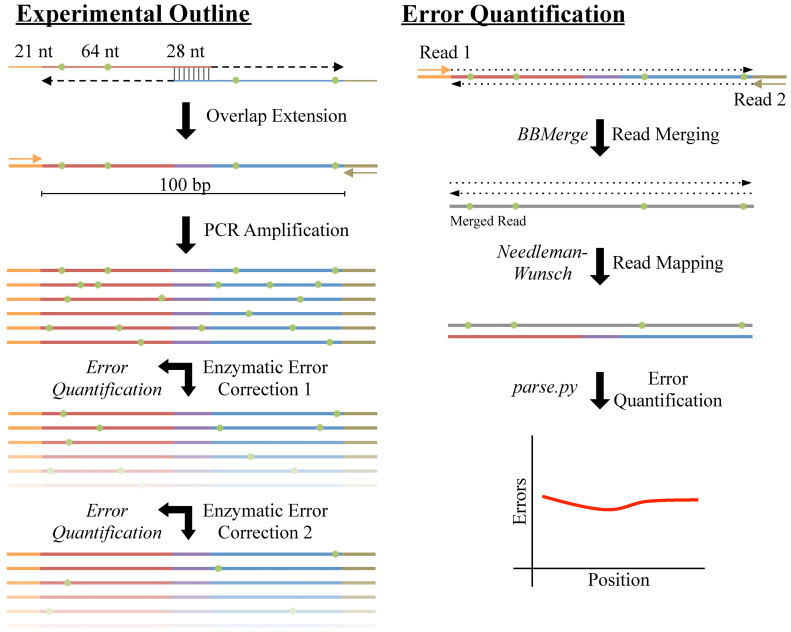
Figure 1.
Schematic of enzymatic error correction and downstream data processing. We assembled our 142 bp product from two 113 nt oligos consisting of a 21 nt primer, a 64 nt payload and a 28 nt overlap region. After annealing and overlap extension, we amplified our template via polymerase chain reaction (PCR), yielding 100 bp of template in-between the primer sites. We then denatured and re-annealed the PCR products to form heteroduplexes, thereby exposing any errors (shown in green). After, we subjected the pool of heteroduplexes to two successive rounds of ten different enzymatic error correction treatments. At each step, we took aliquots and sequenced the products on an Illumina MiSeq with fully overlapping forward and reverse reads. To mitigate sequencing errors, we used BBMerge to merge reads with a perfect agreement between the forward and reverse reads. We then aligned these sequences to the designed reference using an exhaustive Neeleman–Wunsch aligner to minimize alignment artifacts. Finally, we further processed the alignments to quantitate the types and extent of different errors across all conditions.