Abstract
SMADs are essential transcriptional effectors of transforming growth factor-β (TGFβ)/TGFβ-related signaling that underlies embryonic development and adult homeostasis. A recent study by Fang et al. in Cell Research adds to this biological complexity by demonstrating an atypical cytoplasmic role for SMAD5 in modulating the bioenergetic homeostasis (i.e., glycolysis and mitochondrial respiration) of cells in response to fluctuations in intracellular pH that is independent of receptor signaling.
SMADs (homologs of the Drosophila melanogaster protein 'mothers against decapentaplegic' (MAD) and C. elegans 'small body size' (SMA)) are mobile nucleocytoplasmic (phospho) proteins present in all multicellular animals that convey signals produced at the cell surface by the transforming growth factor-β (TGFβ) superfamily of growth factors and cognate receptor serine/threonine kinases into the nucleus where they directly regulate diverse transcriptional programs essential for embryogenesis as well as other key events throughout development1,2. Mammals possess eight distinct SMAD genes (SMAD1-8/9), some of which undergo alternative RNA splicing which further increases their molecular and functional diversity3. While the vast majority of research on SMADs have focused exclusively on their direct nuclear roles in TGF-β-related gene transcription, there is emerging evidence that they may also orchestrate non-genomic events in the absence of receptor activation. This was first described by Kumar et al.4 who demonstrated that 'inactive' (i.e., non-phosphorylated) cytoplasmic SMAD2 associates with the mitochondrial outer membrane where it potently stimulates mitochondrial fusion and energy production via the recruitment of the guanine nucleotide exchange factor 'RAB and RAS Interactor 1' (RIN1) into a complex with mitofusin-2 GTPase (MFN2). This finding raises an intriguing question: Do other SMADs also possess autonomous multitasking capabilities?
The answer appears to be yes. In a recent study by Fang et al.5, the authors provide persuasive evidence that 'inactive' non-phosphorylated SMAD5 also has a cytoplasmic role in energy metabolism. However, in this instance, SMAD5 was found to promote not only mitochondrial respiration but also glycolysis in a manner that was influenced by cytoplasmic pH (pHc) and seemingly distinct from the mitochondrial actions of 'inactive' cytoplasmic SMAD2. The study was initiated by a serendipitous observation that nucleocytoplasmic shuttling of SMAD5 was exquisitely sensitive to acute pHc fluctuations within the physiological range (pHc 7.1 to 7.3) provoked by environmental perturbations of temperature, extracellular pH (pHe) or osmolality. Increases in pHc above 7.2 promoted rapid accumulation of SMAD5 within the cytoplasm, whereas decreases below pHc 7.2 favored its accrual in the nucleus. Significantly, this effect was independent of bone morphogenetic protein (BMP) signaling, SMAD5 C-terminal Ser phosphorylation and interaction with SMAD4, the shared obligate partner of receptor-regulated SMADS (R-SMADs), i.e., a typical pathway for activation of SMAD5-mediated transcriptional events (illustrated in Figure 1A). Moreover, this behavior was relatively specific for SMAD5, as nucleocytoplasmic translocations of other R-SMADs such as SMAD1, SMAD3 and SMAD8 were not affected by pHc fluctuations.
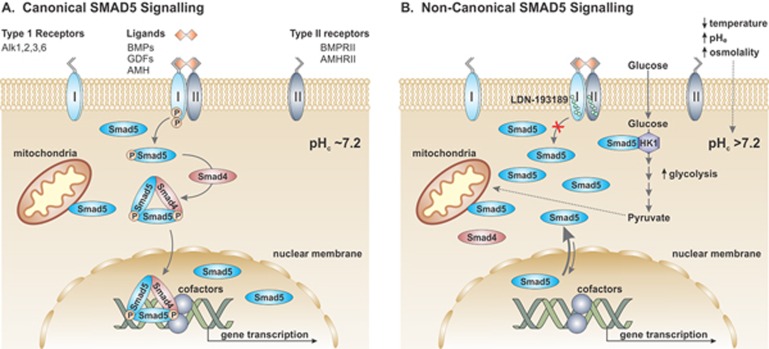
Figure 1.
Canonical and non-canonical SMAD5 signaling. (A) Canonical activation of SMAD5 occurs preferentially upon binding of the TGF-β-related ligands to their cognate receptor kinases. Upon ligand binding, type I and type II receptors form activated, heterotetrameric receptor complexes. Activated type I receptors recruit and phosphorylate SMAD5 on two Ser residues. Two phospho-SMAD5s then assemble as a trimer with SMAD4 and translocate into the nucleus, where they associate with assorted transcription factors and cofactors as well as chromatin-modifying and -remodeling complexes that activate or repress expression of target genes. (B) Non-canonical cytoplasmic signaling of SMAD5 occurs in response to environmental cues (e.g., temperature, pHe and osmolality) and potentially other stimuli that acutely increase cytoplasmic pHc, causing net accumulation of unphosphorylated SMAD5 in the cytoplasm. The more alkaline pHc promotes binding of SMAD5 to HK1, which in turn enhances the rate of glycolysis.
Further mechanistic studies revealed that pHc-sensing by SMAD5 was conferred by one basic (lysine residues) and two acidic (aspartate and glutamate residues) amino acid clusters in its N-terminal MH1 domain. Alanine substitution of the basic residues caused cytoplasmic accumulation of the SMAD5 (SMAD5_K mutant), whereas replacement of the acidic residues (SMAD5-E1E2 mutant) led to its preferential accumulation in the nucleus. Curiously, these charged amino acid clusters in the SMAD5 MH1 domain are also present in pHc-insensitive SMAD1 and SMAD8, indicating that these charged clusters are required but not sufficient for the translocation of SMAD5. The involvement of other residues was further supported by experiments showing that interchanging the MH1 domain of SMAD5 with that of SMAD1, and vice versa, resulted in reciprocal changes in pHc-sensing behavior of the respective chimeric SMADs.
The pH sensitivity of SMAD5 is intriguing in light of recent reports showing that increases in intracellular pH (pHi) are required for the efficient differentiation of embryonic and adult stem cells6. To determine whether there was a functional connection between these seemingly disparate events, the authors performed an elaborate series of experiments that compared the transcriptome profiles of wild-type (WT), SMAD5 knockout (KO) and LDN-193189 (an inhibitor of BMP signaling)-treated human embryonic stem cells (hESCs). The analyses revealed that loss of SMAD5 resulted in the downregulation of a unique subset of genes involved in cellular ion and metabolic homeostasis that were distinct from those affected by blocking BMP signaling. Notably, downregulation of their expression could only be prevented by pre-knockout expression of either the SMAD5 WT or cytoplasmic SMAD5_K mutant, but not the nuclear-localized SMAD5_E1E2 mutant, prior to gene ablation of endogenous SMAD5, thereby implicating a cytoplasmic, non-transcriptional role for SMAD5 in cellular homeostasis.
To more precisely define the cytoplasmic role of SMAD5, the authors examined the metabolic profile of the SMAD5 KO hES cell line and found that these cells exhibited significantly reduced rates of glycolysis and mitochondrial oxygen consumption. Moreover, the mitochondria were visibly swollen and vacuolated and the cell line failed to properly differentiate toward a neural lineage. Strikingly, these deficits could be abrogated by pre-expression of SMAD5 WT or SMAD5_K mutant, but not SMAD5_E1E2. Lastly, co-immunoprecipitation and mass spectrometry experiments identified hexokinase 1 (HK1), a rate limiting enzyme of glycolysis, as a direct binding partner of SMAD5, but not SMAD1 or SMAD8. Significantly, the strength of this interaction increased under more alkaline pHc conditions and correlated with enhanced HK1 activity, both in intact cells and in vitro enzymatic assays. Collectively, these data highlight a novel, non-canonical, cytoplasmic role for SMAD5 as an integrative hub that senses increases in pHi evoked by environmental cues and potentially other stimuli, and translates that signal into enhanced energy production, conditions favorable for proper tissue differentiation (illustrated in Figure 1B).
Lastly, the study by Fang et al. raises new biological questions, perspectives and implications. While the mechanism by which cytoplasmic SMAD5 directly stimulates glycolysis is largely resolved, how it regulates mitochondrial morphology and respiration is less certain. Its ability to modulate respiration could arise indirectly by the increased supply of glycolytically-generated pyruvate to the tricarboxylic acid cycle, but this would not account for its ability to rescue mitochondrial morphology upon SMAD5 ablation. Hints that SMAD5 might have a more direct role in mitochondrial function were provided by an earlier microscopy study7 showing an overlapping distribution between SMAD5 and mitochondria in non-stimulated chondroprogenitor cells, although the nature of that relationship was not determined and merits further investigation. Finally, SMAD5 (along with SMAD2) joins a growing list of 'moonlighting' proteins8 that perform multiple independent and often novel biological functions that deviate markedly from their canonical roles, thus broadening the functional capacity of the human proteome. Whether other SMADs also possess distinct moonlighting functions remains to be determined.
References
- Schmierer B, Hill CS. Nat Rev Mol Cell Biol 2007; 8:970–982. [DOI] [PubMed]
- Massague J. Nat Rev Mol Cell Biol 2012; 13:616–630. [DOI] [PMC free article] [PubMed]
- Tao S, Sampath K. Dev Growth Differ 2010; 52:335–342. [DOI] [PubMed]
- Kumar S, Pan CC, Shah N, et al. Mol Cell 2016; 62:520–531. [DOI] [PMC free article] [PubMed]
- Fang Y, Liu Z, Chen Z, et al. Cell Res 2017; 27:1083–1099. [DOI] [PMC free article] [PubMed]
- Ulmschneider B, Grillo-Hill BK, Benitez M, et al. J Cell Biol 2016; 215:345–355. [DOI] [PMC free article] [PubMed]
- Jullig M, Stott NS. Biochem Biophys Res Commun 2003; 307:108–113. [DOI] [PubMed]
- Huberts DH, van der Klei IJ. Biochim Biophys Acta 2010; 1803:520–525. [DOI] [PubMed]