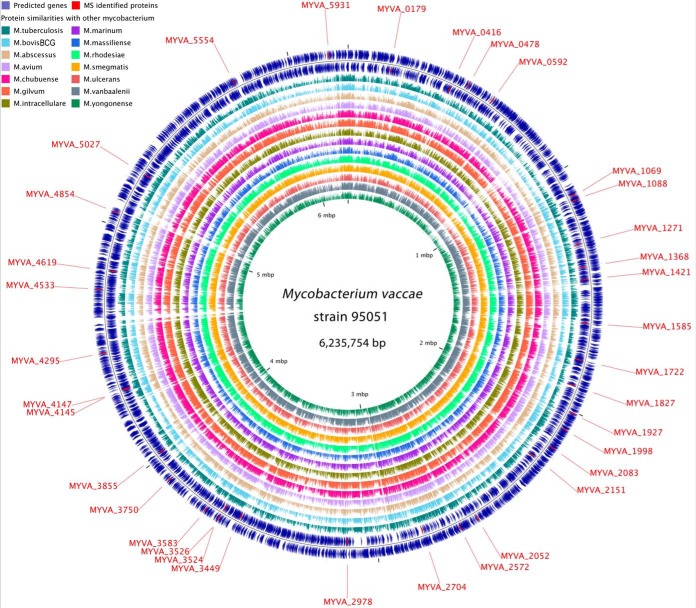
Fig. 1.
Circular genome map of M. vaccae strain 95051. From outer to inner, circles 1 and 2 show the predicted genes on leading and lagging strand, respectively; circles 3 to 16 show genome comparisons in amino acids level with 14 other mycobacterium, including M. tuberculosis H37Rv, M. bovis BCG, M. abscessus ATCC 19977, M. avium strain 104, M. chubuense NBB4, M. gilvum PYR-GCK, M. intracellulare MOTT-02, M. marinum strain M, M. massiliense GO 06, M. rhodesiae NBB3, M. smegmatis MC2 155, M. ulcerans Agy99, M. vanbaalenii PYR-1 and M. yongonense 05–1390. Genome location of the 35 proteins identified by MALDI-TOF/TOF MS are highlighted in red and indicated by outside text. The scale is given on the innermost circle.