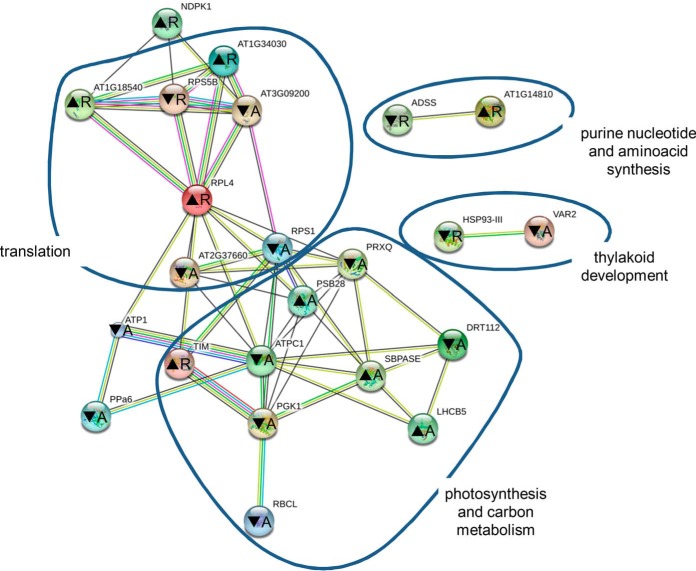
Fig. 3.
Representation of protein interaction networks in differential proteome consistently found in both fra2 and ktn1-2 roots and aerial parts and as generated by STRING web-based application. Letters in nodes mean roots (R) and aerial (A) parts. ▴ means increased abundance, and ▾ means decreased abundance. Different line colors represent the types of evidence used in predicting the associations: gene fusion (red), neighborhood (green), co-occurrence across genomes (blue), co-expression (black), experimental (purple), association in curated databases (light blue), or co-mentioned in PubMed abstracts (yellow). RPL4 is 50S ribosomal protein L4 (gi79317147); At1g14810 is aspartate semialdehyde dehydrogenase (gi15223910); ADSS is adenylosuccinate synthetase (gi15230358); At1g18540 is 60S ribosomal protein L6-1 (gi15221798); DRT112 is DNA-damage resistance protein DRT112 (gi15217918); At1g34030 is 40S ribosomal protein S18 (gi18399100); TIM is triose-phosphate isomerase (gi145329204); VAR2 is ATP-dependent zinc metalloprotease FTSH 2 (VAR2) (gi30684767); RPS5B is 40S ribosomal protein S5-1 (gi79324564); At2g37660 is NAD(P)-binding Rossmann fold-containing protein (gi18404496); At3g09200 is 60S acidic ribosomal protein P0-2 (gi15232603); PGK1 is phosphoglycerate kinase 1 (gi15230595); PRXQ is peroxiredoxin Q (gi15230982); HSP93-III is Clp ATPase (gi334185828); SBPASE is sedoheptulose-1,7-bisphosphatase (gi15228194); ATPC1 is ATP synthase γ chain 1 (gi18412632); NDPK1 is nucleoside diphosphate kinase 1 (gi18413214); LHCB5 is chlorophyll a-b-binding protein CP26 (gi15235029); PSB28 is photosystem II reaction center PSB28 protein (gi18417239); PPa6 is soluble inorganic pyrophosphatase 1 (gi15242465); RPS1 is small subunit ribosomal protein S1 (gi30692346); RBCL is ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (gi7525041); and ATP1 is ATPase subunit 1 (gi26557005).