Abstract
Pancreatic cancer, one of the most common cancers globally, is the fourth most common cause of cancer-associated mortality in the USA. The 5-year relative survival rate for patients with pancreatic cancer is ~5% and the median survival time is only 6 months. The poor prognosis is mainly due to early and aggressive local invasion and metastasis, as well as dissemination of the pancreatic cancer cells. The present study demonstrated that microRNA-452 (miR-452) was markedly downregulated in pancreatic cancer tissues, particularly in metastatic tumors and pancreatic cancer cell lines. Overexpression of miR-452 significantly inhibited migration and invasion in pancreatic cancer cells. In addition, the molecular mechanism underlying the inhibitory functions of miR-452 in pancreatic cancer was also investigated. The results indicated that B-cell-specific Moloney murine leukemia virus insertion site 1 (BMI1) was a direct target gene of miR-452 in pancreatic cancer. Overexpression of miR-452 inhibited the migration and invasion of pancreatic cancer, at least partially by knockdown of BMI1 expression. The results provided novel insight with potential therapeutic applications for the treatment of metastatic pancreatic cancer.
Keywords: pancreatic cancer, pancreatic ductal adenocarcinoma, microRNA-452, B-cell-specific Moloney murine leukemia virus insertion site 1, metastasis, therapy
Introduction
Pancreatic cancer, one of the most common cancers globally, is the fourth most common cause of cancer-associated mortality in the USA (1). In 2015, it was estimated that there would be 48,960 new pancreatic cancer cases and 40,560 mortalities due to pancreatic cancer in the USA (2). Epidemiological studies have demonstrated that the likelihood of pancreatic cancer is associated with numerous factors, including smoking, high-fat high-protein diets, alcoholism, coffee drinking, exposure to certain chemical carcinogens, diabetes mellitus and chronic pancreatitis (3,4). Pancreatic ductal adenocarcinoma is the most common subtype of pancreatic cancer, accounting for 85–90% of all pancreatic cancer cases (5). Although progress has been made in surgery and perioperative management, the 5-year relative survival rate for patients with pancreatic cancer remains at ~5% and the median survival time is only 6 months (6). The poor prognosis is mainly a result of early and aggressive local invasion and metastasis, as well as dissemination of the pancreatic cancer cells (7). Therefore, it is urgent to improve understanding of the molecular mechanisms underlying the metastasis of pancreatic cancer, and to investigate more effective therapeutic treatments for patients with pancreatic cancer to block cancer metastasis.
MicroRNAs (miRNAs/miRs) are considered to be useful biomarkers for the early diagnosis, therapy and prognosis of human cancers, including pancreatic cancer (8–10). They are an evolutionarily conserved group of non-protein-coding and short RNAs, 18–22 nucleotides in length, whose function is to regulate the expression of their target protein-coding genes either by translational suppression or gene degradation through binding to the 3′ untranslated regions (3′UTRs) of target genes in a base-pairing manner (11,12). An increasing number of studies have demonstrated that miRNAs regulate numerous biological processes, including cell growth, apoptosis, the cell cycle, metastasis and metabolism, and consequently, their alterations are considered to perform important functions in carcinogenesis and progression of cancers (13). Abnormal expression of miRNAs is associated with various types of diseases, including cancer, and tumor-associated miRNAs act as tumor suppressors or oncogenes depending on their target mRNAs (14). Therefore, elucidating the expression, function and molecular mechanism of miRNAs will be particularly useful in investigating novel therapeutic treatments for cancers.
miR-452 has been studied in several types of cancer, including non-small cell lung cancer (15), breast cancer (16), bladder cancer (17) and hepatocellular carcinoma (18). However, the expression, biological function and molecular mechanism of miR-452 in pancreatic cancer remains to be fully elucidated. The present study revealed that miR-452 was significantly downregulated in pancreatic cancer tissues, particularly in metastatic tumors and pancreatic cancer cell lines. Notably, migration and invasion assays indicated that overexpression of miR-452 decreased the migration and invasion of pancreatic cancer cells. Subsequent experiments demonstrated that BMI1 was a direct target gene of miR-452 in pancreatic cancer. Collectively, miR-452 suppressed the migration and invasion of pancreatic cancer cells by directly targeting BMI1.
Materials and methods
Human tissue specimens and ethics statement
A total of 32 pancreatic cancer tissues with matched adjacent normal tissues were obtained from patients (male, 19; female, 13; age, 46–69 years; mean, 57 years) during surgery at Weifang People's Hospital (Weifang, China) between June 2013 and March 2015. None of patients underwent chemotherapy or radiotherapy prior to surgery. All the tissues were immediately snap-frozen and stored in liquid nitrogen until use. For the analyzed tissue specimens, written informed consent was obtained from all patients to use excess pathological specimens for study purposes. The present study was approved by the Ethics Committee of Weifang People's Hospital and performed according to the ethical principles.
Cell culture and cell transfection
The human pancreatic cancer cell lines (PANC-1, SW1990, ASPC-1 and CFPAC-1) and the human normal pancreatic cell line (HPDE6c7) were obtained from American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 IU/ml penicillin and 100 µg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cell lines were cultured in a humidified cell incubator, with an atmosphere of 5% CO2 and 95% air, at 37°C.
miR-452 mimic and negative control (NC) were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). BMI1 small interfering RNA (siRNA) and NC siRNA were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The miR-452 mimic sequence was 5′-AACUGUUUGCAGAGGAAACUGA-3′ and the NC sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The BMI1 siRNA sequence was 5′-GUUCACAAGACCAGACCAC-3′ and the NC siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. miRNA or siRNA were transfected into cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocol. The medium was replaced with fresh culture medium 6–8 h after transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues and cell lines using TRIzol regent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. For miRNA expression, RT-qPCR assays were performed using a TaqMan miRNA Assay in an Applied Biosystems 7500 Real-time PCR system (both from Applied Biosystems; Thermo Fisher Scientific, Inc.). For mRNA expression, cDNA was generated from total RNA through reverse transcription using M-MLV reverse transcriptase (Promega Corporation, Madison, WI, USA). BMI1 mRNA expression was evaluated with SYBR-Green Master Mix (Takara Biotechnology Co., Ltd., Dalian, China). The thermocycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primers were designed as follows: miR-452, 5′-GCGAACTGTTTGCAGAGG-3′ (forward) and 5′-CAGTGCGTGTCGTGGAGT-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-ACGCTTCACCGAATTTGAGT-3′ (reverse); BMI1, 5′-GTGCTTTGTGGAGGGTACTTCAT-3′ (forward) and 5′-TTGGACATCACAAATAGGACAATACTT-3′ (reverse); and GAPDH, 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGGGATTTC-3′ (reverse). Each sample was analyzed in triplicate. U6 and GADPH were used as internal controls for miR-452 and BMI1 mRNA expression, respectively. Relative expression levels were calculated using the 2−∆∆Cq method (19).
Migration and invasion assay
Transwell chambers (Costar; Corning Incorporated, Corning, NY, USA) with 8-mm pore size polycarbonate membranes were applied to examine cell migration and invasion capacity. For the invasion assay, Transwell chambers were coated with Matrigel (40 µg/well; BD Biosciences, San Jose, CA, USA) prior to the experiment. In brief, 1×105 transfected cells suspended in 200 µl serum-free medium were placed in the top chambers and 500 µl complete culture medium containing 10% FBS was added to the lower chambers.
Following incubation at 37°C in a cell incubator for 48 h, the cells on the top surface of the Transwell chamber were gently removed from the top chambers with a cotton swab. Cells on the lower surface of the membrane were then fixed with 90% methanol for 10 min and stained with 0.1% crystal violet for 10 min, and averaged across five random fields at ×100 magnification using an inverted microscope (Olympus Corporation, Tokyo, Japan).
Target prediction of miRNAs
TargetScan (http://www.targetscan.org/) was adopted to identify the potential targets of miR-452.
Western blot analysis
Following incubation at 37°C for 72 h post-transfection, cells were lysed using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) supplemented with 0.1 mg/ml phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate and 1 mg/ml aprotinin. Protein concentration was determined using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology). Equal amounts of protein (20 mg) were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA) using a Bio-Rad semidry transfer system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membranes were then blocked in 5% skim milk in TBS-Tween-20 (TBST) for 2 h at room temperature, followed by incubation with the following primary antibodies: Mouse anti-human monoclonal BMI1 (1:1,000 dilution; cat no. sc-13519) and mouse anti-human GADPH (1:1,000 dilution; cat no. sc-166574) (both from Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. The membranes were washed three times with TBST and probed with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; cat no. sc-2005; Santa Cruz Biotechnology, Inc.) at room temperature for 1 h. Membranes were then washed three times with TBST and visualized with enhanced chemiluminescence plus reagents (Pierce; Thermo Fisher Scientific, Inc.). Protein expression was quantified using Quantity One software version 4.62 (Bio-Rad Laboratories, Inc.). GADPH was used as an internal control for BMI1 protein expression.
Dual-luciferase reporter assay
PGL3-BMI1-3′UTR wild-type (Wt) and PGL3-BMI1-3′UTR mutant (Mut) were purchased from Shanghai GenePharma Co., Ltd. Cells were seeded onto 96-well plates and transfected with miR-452 mimics or NC, and co-transfected with PGL3-BMI1-3′UTR Wt or PGL3-BMI1-3′UTR Mut using Lipofectamine 2000 according to the manufacturer's protocol. Following incubation at 37°C for 48 h, cells were collected and luciferase activities were measured using a Dual-Luciferase Reporter Assay system (Promega Corporation). The relative luciferase activities were normalized to that of Renilla activity.
Statistical analysis
Data are expressed as the mean ± standard deviation, and compared using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Groups were compared using two-tailed Student's t-tests or one-way analysis of variance with Student-Newman-Keuls post hoc tests. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-452 is frequently downregulated in pancreatic cancer tissues and cell lines
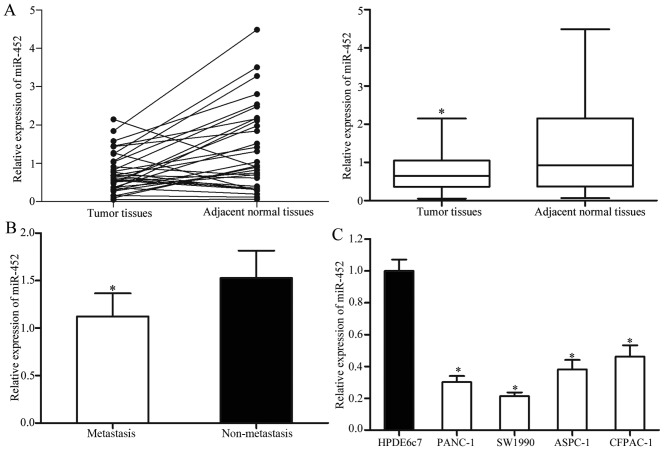
The expression levels of mature miR-452 in the pancreatic cancer tissues and matched adjacent normal tissues were measured using RT-qPCR. The results revealed that miR-452 was significantly decreased in the pancreatic cancer tissues compared with that in matched adjacent normal tissues (Fig. 1A). In addition, the levels of miR-452 in metastatic pancreatic cancer tissues were significantly lower compared with that in pancreatic cancer without metastasis (Fig. 1B).
Figure 1.
miR-452 expression levels in pancreatic cancer tissues and cell lines. (A) Expression levels of miR-452 in pancreatic cancer tissues and matched adjacent normal tissues. (B) Levels of miR-452 in metastatic pancreatic cancer tissues and pancreatic cancer without metastasis. (C) miR-452 expression levels in four pancreatic cancer cell lines and the human normal pancreatic cell line. *P<0.05 vs. the respective controls. miR-452, microRNA-452.
miR-452 expression levels were also explored in four pancreatic cancer cell lines (PANC-1, SW1990, ASPC-1 and CFPAC-1) and the human normal pancreatic cell line (HPDE6c7). Lower expression levels of miR-452 were observed in all four pancreatic cancer cell lines compared with that in the HPDE6c7 cell line (Fig. 1C). Among these cell lines, PANC-1 and SW1990 cells were selected for subsequent functional studies as they expressed relatively lower levels of miR-452.
Overexpression of miR-452 inhibits pancreatic cancer migration and invasion
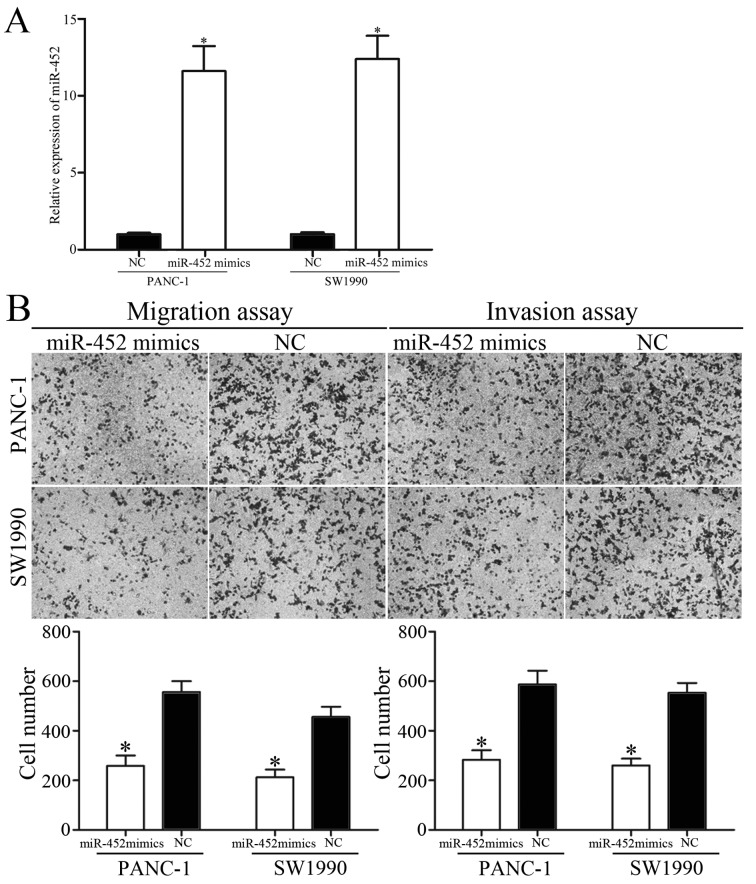
Since miR-452 was downregulated in patients with metastatic pancreatic cancer, whether overexpression of miR-452 decreased migration and invasion capacity of pancreatic cancer was subsequently explored. PANC-1 and SW1990 cells were transfected with miR-452 mimics or NC. The ectopic expression efficiency of miR-452 was validated by RT-qPCR (Fig. 2A).
Figure 2.
Overexpression of miR-452 inhibited pancreatic cancer migration and invasion. (A) Expression levels of miR-452 in PANC-1 and SW1990 cells. PANC-1 and SW1990 cells were transfected with miR-452 mimics or NC. (B) Cell migration and invasion capacity of PANC-1 and SW1990 cells was assessed by migration and invasion assays following transfection with miR-452 mimics or NC. *P<0.05 vs. the respective controls. miR-452, microRNA-452; NC, negative control.
To clarify the effect of miR-452 on tumor metastasis, migration and invasion assays were performed with Transwell chambers. The results revealed that the migratory and invasive abilities of PANC-1 and SW1990 cells were suppressed by miR-452 overexpression (Fig. 2B). Collectively, these results indicated that miR-452 functioned as a tumor suppressor and contributed to suppression of metastasis of pancreatic cancer.
BMI1 is a direct target gene of miR-452 in pancreatic cancer
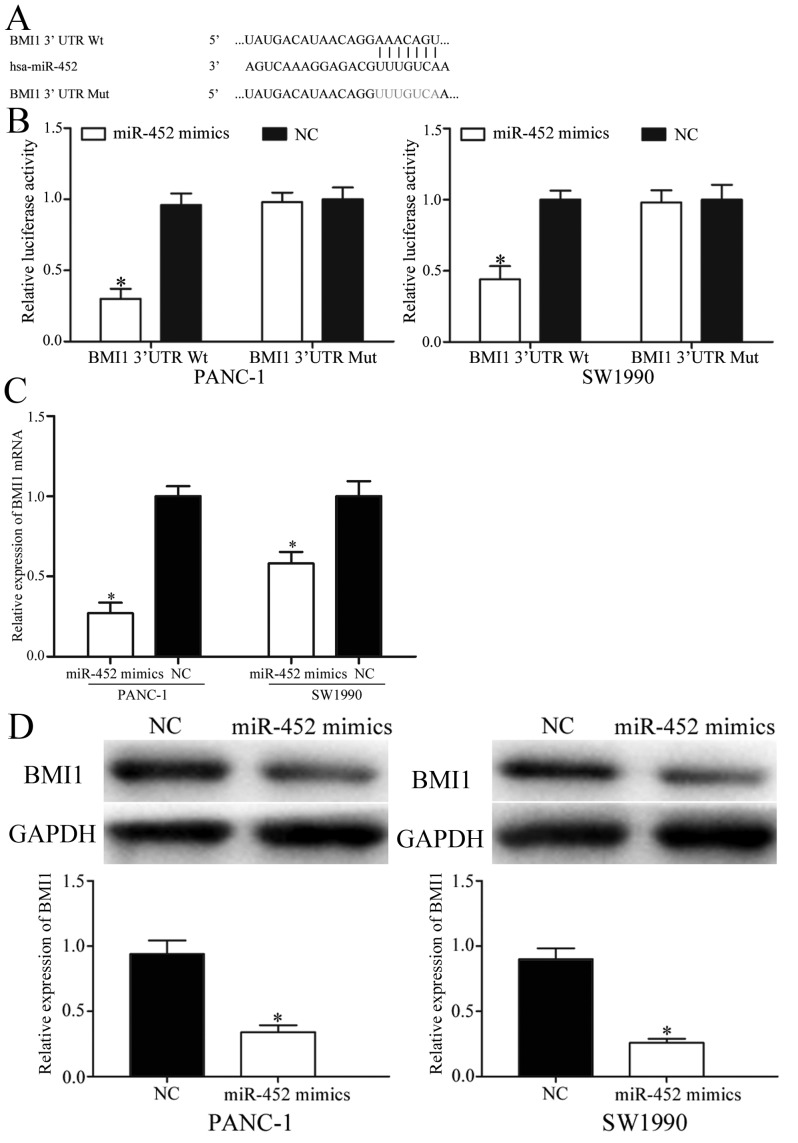
To further elucidate the molecular mechanisms underlying the inhibitory functions of miR-452 in pancreatic cancer, TargetScan was applied to identify predicated targets. As presented in Fig. 3A, BMI1 contained the predicted binding sites for miR-452. Dual-luciferase reporter assays were then performed to investigate whether miR-452 directly targeted the 3′UTR of BMI1. The results revealed that co-transfection of miR-452 significantly decreased the activities of the firefly luciferase reporter with BMI1-3′UTR Wt, whereas the inhibition effect was abolished when the predicted 3′UTR binding sites of BMI1 were mutated (Fig. 3B).
Figure 3.
BMI1 was a direct target of miR-452 in pancreatic cancer. (A) The predicted binding sequences for BMI1 3′UTR with miR-452. (B) Dual-luciferase reporter assays were used to measure the interaction between miR-452 and the BMI1 3′UTR. The BMI1 3′UTR Wt or BMI1 3′UTR Mut was co-transfected with miR-452 mimics or NC and incubated for 48 h with PANC-1 and SW1990 cells. (C) The mRNA levels of BMI1 were detected by reverse transcription-quantitative polymerase chain reaction. PANC-1 and SW1990 cells were transfected with miR-452 mimics or NC. (D) BMI1 protein levels were measured by western blot analysis. PANC-1 and SW1990 cells were transfected with miR-452 mimics or NC. *P<0.05 vs. the respective controls. BMI1, B-cell-specific Moloney murine leukemia virus insertion site 1; miR-452, microRNA-452; 3′UTR, 3′ untranslated region; NC, negative control; Wt, wild-type; Mut, mutant.
Furthermore, RT-qPCR and western blot analysis were adopted to determine the regulatory effects of miR-452 in BMI1 expression. The results indicated that overexpression of miR-452 markedly suppressed BMI1 mRNA and protein levels in PANC-1 and SW1990 cells (Fig. 3C and D). Taken together, BMI1 was a direct target gene of miR-452 in pancreatic cancer.
Overexpression of miR-452 inhibits migration and invasion of pancreatic cancer cells via knockdown of BMI1
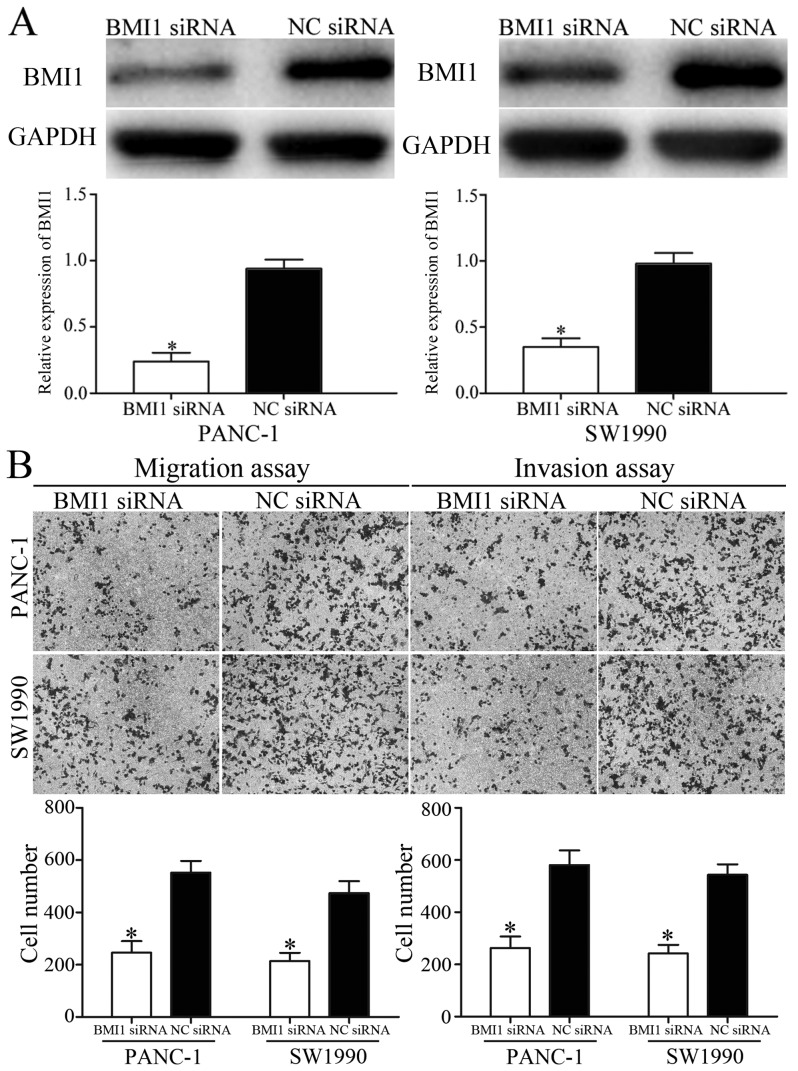
BMI1 was verified as a direct target gene of miR-452 in pancreatic cancer. To explore whether the inhibition effects of miR-452 on migration and invasion of pancreatic cancer was achievable by knockdown of BMI1, BMI1 siRNA or NC siRNA were transfected into PANC-1 and SW1990 cells. The transfection efficiency was measured using western blot analysis (Fig. 4A). The effect of BMI1 siRNA in metastasis of pancreatic cancer was also measured. The migration and invasion assays indicated that BMI1 siRNA inhibited migration and invasion capacity of PANC-1 and SW1990 cells compared with that in NC siRNA groups (Fig. 4B). These results suggested that overexpression of miR-452 inhibited migration and invasion of pancreatic cancer, at least partially by knockdown of BMI1 expression.
Figure 4.
Knockdown of BMI1 inhibited pancreatic cancer migration and invasion. (A) BMI1 protein levels were measured by western blot analysis. PANC-1 and SW1990 cells were transfected with BMI1 siRNA or NC siRNA. (B) Cell migration and invasion of PANC-1 and SW1990 cells was assessed using Transwell migration and invasion assays following transfection with BMI1 siRNA or NC siRNA. *P<0.05 vs. the respective controls. BMI1, B-cell-specific Moloney murine leukemia virus insertion site 1; siRNA, small interfering RNA; NC, negative control.
Discussion
Pancreatic cancer is an aggressive tumor characterized by early and aggressive metastasis, as well as dissemination of pancreatic cancer (7). Successful management and therapy of patients with pancreatic cancer remains one of the major challenges in clinical oncology. It is possible to cure patients with early stage pancreatic cancer with surgical operation (20). However, the majority of patients are diagnosed at advanced stages and surgery is not possible (21). Therefore, there is an urgent requirement for novel therapeutic treatments for patients with pancreatic cancer. A number of studies have indicated that miRNAs contribute to the carcinogenesis and progression of numerous types of human cancer via targeting multiple mRNAs (22–24). Therefore, identification of specific miRNAs may provide therapeutic implications and may be exploited to improve treatments for patients with pancreatic cancer.
The present study is the first to investigate the expression, biological functions and molecular mechanism of miR-452 in pancreatic cancer. It was demonstrated that miR-452 was significantly downregulated in pancreatic cancer tissues and cell lines. In addition, the levels of miR-452 in metastatic pancreatic cancer tissues were significantly lower compared with those in pancreatic cancer without metastasis. Functional studies indicated that overexpression of miR-452 suppressed the migration and invasion of pancreatic cancer. In addition, BMI1 was identified as a direct target gene of miR-452 in pancreatic cancer. Thus, miR-452 acted as a tumor suppressor in pancreatic cancer and its downregulation enhanced metastasis of pancreatic cancer.
miR-452 has been studied in numerous types of human cancer (15,16,25). For example, He et al (15) reported that the expression levels of miR-452 were decreased in non-small-cell lung cancer, and low expression levels of miR-452 were associated with advanced tumor stage and more extent of lymph nodes metastasis. Functional studies have revealed that under-expression of miR-452 may induce cell invasion of non-small-cell lung cancer (15). In addition, miR-452 was revealed to negatively regulate BMI1 expression by binding to its 3′UTR, and the enhanced cell invasion induced by downregulated miR-452 may be eliminated by knockdown of BMI1 (15). In breast cancer, miR-452 was downregulated in adriamycin-resistant MCF-7 cells compared with in parental MCF-7 cells. miR-452 mediated chemosensitivity to adriamycin in human breast cancer cells (16). Furthermore, bioinformatics analysis, RT-qPCR and western blot analysis have demonstrated that miR-452 may be at least in part be involved in adriamycin-resistance of breast cancer via the targeting of insulin-like growth factor-1 (16). Liu et al (25) revealed that miR-452 was significantly downregulated in glioma tissues and cell lines, and miR-452 levels were associated with World Health Organization grades and patient survival. Additional studies revealed that miR-452 targeted BMI1, lymphoid enhancer-binding factor 1 and T cell factor 4 to decrease glioma stem-like phenotypes in vitro and inhibit glioma carcinogenesis in vivo (21).
miR-452 is regarded as a tumor suppressor in non-small cell lung cancer (15), breast cancer (16) and glioma (25). However, in hepatocellular carcinoma, miR-452 was markedly upregulated in tumor tissues and cell lines (18). Ectopic expression of miR-452 enhanced cell proliferation, promoted transition from phase G1 to S in the cell cycle and suppressed apoptosis, migration and invasion of hepatocellular carcinoma. In addition, miR-452 decreased cyclin-dependent kinase inhibitor 1B (CDKN1B) mRNA and protein expression by directly targeting the 3′UTR of CDKN1B; the knockdown of CDKN1B resembled the phenotype resulting from overexpression of miR-452 expression (18). miR-452 was also demonstrated to be an oncogene in esophageal cancer (26) and urothelial carcinoma (27). These conflicting studies indicated that the expression and functions of miR-452 in human cancers have tissue specificity. It may be explained by distinct context of various microenvironments, and the imperfect complementarity of the interactions between miRNAs and target mRNAs (28). In the present study, miR-452 was demonstrated to act as a tumor suppressor in pancreatic cancer by inhibiting cell migration and invasion. The present study expanded the expression and functions of miR-452 in human cancers.
Identifying the target genes of miRNA is essential for understanding its functions in carcinogenesis and progression. In the present study, BMI1 was revealed to be the direct target gene of miR-452 in pancreatic cancer. BMI1, a member of the Polycomb group genes, was first demonstrated as an oncogene in murine lymphoma (29). Previous studies have suggested that BMI1 is highly expressed in numerous human cancers, including lung cancer (30), prostate cancer (31) and colorectal cancer (32). Additional studies have demonstrated that BMI1 serves important functions in a number of biological processes, including the cell cycle, apoptosis, senescence and proliferation (33–35). In pancreatic cancer, BMI1 is upregulated and expression levels of BMI1 are associated with proliferation, survival and poor prognosis of patients with pancreatic cancer (36). Furthermore, BMI1 functioned as an oncogene in pancreatic cancer by promoting chemoresistance, invasion and tumorigenesis of pancreatic cancer (37). The results of the present study were in accordance with the aforementioned results. The present study revealed that knockdown of BMI1 suppressed migration and invasion of pancreatic cancer cells, which is similar to the effects of miR-452 in pancreatic cancer.
In summary, the present study demonstrated that miR-452 was downregulated in pancreatic cancer tissues, particularly in metastatic tumors and pancreatic cancer cell lines. Ectopic expression of miR452 may suppress migration and invasion of pancreatic cancer cells through directly targeting BMI1. These results indicated that miR-452 may be useful as a novel potential therapeutic treatment for pancreatic cancer to block metastasis.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Moir J, White SA, French JJ, Littler P, Manas DM. Systematic review of irreversible electroporation in the treatment of advanced pancreatic cancer. Eur J Surg Oncol. 2014;40:1598–1604. doi: 10.1016/j.ejso.2014.08.480. [DOI] [PubMed] [Google Scholar]
- 4.Burkey MD, Feirman S, Wang H, Choudhury SR, Grover S, Johnston FM. The association between smokeless tobacco use and pancreatic adenocarcinoma: A systematic review. Cancer Epidemiol. 2014;38:647–653. doi: 10.1016/j.canep.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ, Ou JR. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 2015;589:2224–2232. doi: 10.1016/j.febslet.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Huang C, Xie K. Analysis of the potential for pancreatic cancer metastasis in vitro and in vivo. Methods Mol Biol. 2013;980:301–319. doi: 10.1007/978-1-62703-287-2_17. [DOI] [PubMed] [Google Scholar]
- 7.Su D, Yamaguchi K, Tanaka M. The characteristics of disseminated tumor cells in pancreatic cancer: A black box needs to be explored. Pancreatology. 2005;5:316–324. doi: 10.1159/000086532. [DOI] [PubMed] [Google Scholar]
- 8.Qin Y, Dang X, Li W, Ma Q. miR-133a functions as a tumor suppressor and directly targets FSCN1 in pancreatic cancer. Oncol Res. 2013;21:353–363. doi: 10.3727/096504014X14024160459122. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Ou Y, Wu K, Chen Y, Sun W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biol. 2012;33:1863–1870. doi: 10.1007/s13277-012-0446-8. [DOI] [PubMed] [Google Scholar]
- 10.Tréhoux S, Lahdaoui F, Delpu Y, Renaud F, Leteurtre E, Torrisani J, Jonckheer N, Van Seuningen I. Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells. Biochim Biophys Acta. 2015;1853:2392–2403. doi: 10.1016/j.bbamcr.2015.05.033. [DOI] [PubMed] [Google Scholar]
- 11.Guled M, Knuutila S. MicroRNAs and cancer. Duodecim. 2013;129:1661–1669. (In Finnish) [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z, Xia Y, Pan C, Ma T, Liu B, Wang J, Chen L, Chen Y. Up-regulation of miR-452 inhibits metastasis of non-small cell lung cancer by regulating BMI1. Cell Physiol Biochem. 2015;37:387–398. doi: 10.1159/000430362. [DOI] [PubMed] [Google Scholar]
- 16.Hu Q, Gong JP, Li J, Zhong SL, Chen WX, Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH, Tang JH. Down-regulation of miRNA-452 is associated with adriamycin-resistance in breast cancer cells. Asian Pac J Cancer Prev. 2014;15:5137–5142. doi: 10.7314/APJCP.2014.15.13.5137. [DOI] [PubMed] [Google Scholar]
- 17.Puerta-Gil P, Garcia-Baquero R, Jia AY, Ocaña S, Alvarez-Múgica M, Alvarez-Ossorio JL, Cordon-Cardo C, Cava F, Sánchez-Carbayo M. miR-143, miR-222, and miR-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer. Am J Pathol. 2012;180:1808–1815. doi: 10.1016/j.ajpath.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Q, Sheng Q, Jiang C, Shu J, Chen J, Nie Z, Lv Z, Zhang Y. MicroRNA-452 promotes tumorigenesis in hepatocellular carcinoma by targeting cyclin-dependent kinase inhibitor 1B. Mol Cell Biochem. 2014;389:187–195. doi: 10.1007/s11010-013-1940-z. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Link KH, Leder G, Formentini A, Fortnagel G, Kornmann M, Schatz M, Beger HG. Surgery and multimodal treatments in pancreatic cancer-a review on the basis of future multimodal treatment concepts. Gan To Kagaku Ryoho. 1999;26:10–40. [PubMed] [Google Scholar]
- 21.Vychytilova-Faltejskova P, Kiss I, Klusova S, Hlavsa J, Prochazka V, Kala Z, Mazanec J, Hausnerova J, Kren L, Hermanova M, et al. miR-21, miR-34a, miR-198 and miR-217 as diagnostic and prognostic biomarkers for chronic pancreatitis and pancreatic ductal adenocarcinoma. Diagn Pathol. 2015;10:38. doi: 10.1186/s13000-015-0272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 23.Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62:33–40. doi: 10.1038/jhg.2016.59. [DOI] [PubMed] [Google Scholar]
- 24.Zhu M, Xu Z, Wang K, Wang N, Li Y. microRNA and gene networks in human pancreatic cancer. Oncol Lett. 2013;6:1133–1139. doi: 10.3892/ol.2013.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Chen K, Wu J, Shi L, Hu B, Cheng S, Li M, Song L. Downregulation of miR-452 promotes stem-like traits and tumorigenicity of gliomas. Clin Cancer Res. 2013;19:3429–3438. doi: 10.1158/1078-0432.CCR-12-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ, Wang TY, Li HC, Wu XN. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett. 2013;5:1639–1642. doi: 10.3892/ol.2013.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, Liedberg F, Chebil G, Gudjonsson S, Borg A, et al. miRNA expression in urothelial carcinomas: Important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236–2242. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 28.Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y, Yu W, Wu X, Ye J, Yang S, et al. Identification of miR-7 as an oncogene in renal cell carcinoma. J Mol Histol. 2013;44:669–677. doi: 10.1007/s10735-013-9516-5. [DOI] [PubMed] [Google Scholar]
- 29.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 30.Xiong D, Ye Y, Fu Y, Wang J, Kuang B, Wang H, Wang X, Zu L, Xiao G, Hao M, Wang J. Bmi-1 expression modulates non-small cell lung cancer progression. Cancer Biol Ther. 2015;16:756–763. doi: 10.1080/15384047.2015.1026472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolters T, Vissers KJ, Bangma CH, Schröder FH, van Leenders GJ. The value of EZH2, p27(kip1), BMI-1 and MIB-1 on biopsy specimens with low-risk prostate cancer in selecting men with significant prostate cancer at prostatectomy. BJU Int. 2010;106:280–286. doi: 10.1111/j.1464-410X.2009.08998.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Yang X, Zhang Y, Liu X, Zheng G, Yang Y, Wang L, Du L, Wang C. Direct serum assay for cell-free bmi-1 mRNA and its potential diagnostic and prognostic value for colorectal cancer. Clin Cancer Res. 2015;21:1225–1233. doi: 10.1158/1078-0432.CCR-14-1761. [DOI] [PubMed] [Google Scholar]
- 33.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI200420800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K, Adhikary G, Balasubramanian S, Gopalakrishnan R, McCormick T, Dimri GP, Eckert RL, Rorke EA. Expression of Bmi-1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J Invest Dermatol. 2008;128:9–17. doi: 10.1038/sj.jid.5700949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasini D, Bracken AP, Helin K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle. 2004;3:396–400. doi: 10.4161/cc.3.4.773. [DOI] [PubMed] [Google Scholar]
- 36.Song W, Tao K, Li H, Jin C, Song Z, Li J, Shi H, Li X, Dang Z, Dou K. Bmi-1 is related to proliferation, survival and poor prognosis in pancreatic cancer. Cancer Sci. 2010;101:1754–1760. doi: 10.1111/j.1349-7006.2010.01577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin T, Wei H, Leng Z, Yang Z, Gou S, Wu H, Zhao G, Hu X, Wang C. Bmi-1 promotes the chemoresistance, invasion and tumorigenesis of pancreatic cancer cells. Chemotherapy. 2011;57:488–496. doi: 10.1159/000334103. [DOI] [PubMed] [Google Scholar]