Figure 2. Protecting groups can be coupled to amino acids adjacent to scissile bond.
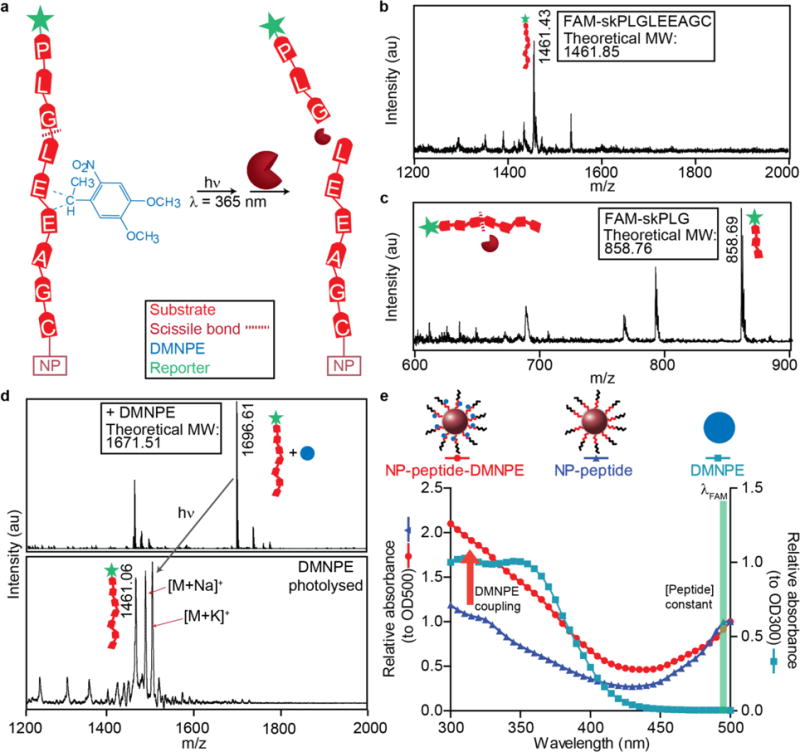
(a) The peptide backbone can be directly modified with a photolabile group (DMNPE; blue) at acidic residues. Adjacent to the peptide substrate, reporters that can be either fluorigenic or ligand-encoded (green) are released up cleavage. Activation by light removes the photolabile group and enables proteases to access the peptide. (b) Mass spectrometry analysis of the native peptide sequence. (c) Identification of the scissile bond by mass spectrometry analysis of MMP9 cleaved peptide fragment. (d) Coupling of a DMNPE molecule is confirmed by an m/z shift corresponding to the mass of one DMNPE molecule. Photolysis results in a mass shift back to the original mass of the native peptide. (e) Spectral characteristic of NP-peptides (triangles) and spectral shift with DMNPE coupled (circle) that approximately matches spectra of free DMNPE.
