Abstract
Fluorescent in situ hybridization (FISH) is a technique for determining the cytological localization of RNA or DNA molecules. There are many approaches available for generating in situ hybridization probes and conducting the subsequent hybridization steps. Here, we describe a simple and reliable FISH method to label small RNAs (200–500 nucleotides in length) that are enriched in nuclear bodies in Drosophila melanogaster ovaries, such as Cajal bodies (CBs) and histone locus bodies (HLBs). This technique can also be applied to other Drosophila tissues, and to abundant mRNAs such as histone transcripts.
Keywords: Fluorescence in situ hybridization (FISH), Small nuclear RNAs (snRNAs), Small Cajal body-specific RNAs (scaRNAs), Cajal bodies (CBs), Histone locus bodies (HLBs), Drosophila ovaries
1 Introduction
The nucleus of a cell is organized into non membrane-bound compartments such as CBs and HLBs, which are enriched in factors involved in pre-mRNA processing [1]. These bodies can be identified based on their molecular composition, either by antibody staining to label proteins or in situ hybridization to label RNAs. When labeling nuclear bodies, it is important to examine more than one marker because there can be situations when a particular protein or RNA is simultaneously enriched in two or more distinct classes of nuclear bodies. For example, coilin is a protein that has been widely used as a molecular marker of CBs, but in certain Drosophila tissues and at various stages of oogenesis, it is also enriched in HLBs [2]. As such, there is presently no one antibody that robustly and reliably acts as a unique marker of Drosophila CBs without some prior characterization of the tissue. In contrast, there are small RNAs in these bodies (200–500 nucleotides in length) that are uniquely localized. These include small Cajal body-specific RNAs (scaRNAs) and spliceosomal U small nuclear RNAs (snRNAs) that are enriched in CBs, and the U7 snRNA that is enriched in HLBs [3].
To label these small RNAs in CBs and HLBs, we use the simple and reliable FISH method described here. The simplicity lies in the generation of directly labeled fluorescent probes, which enables one to skip downstream labeling and amplification steps and proceed directly from hybridization to mounting the specimen. The reliability of the method stems from the cytological concentration of the target RNAs in foci such as CBs and HLBs.
In this protocol, we detail the specific methodology for FISH to RNA targets in CBs and HLBs in whole mount Drosophila ovaries [3] (Fig. 1). This protocol may be applied without modification to other Drosophila tissues and tissues from other organisms such as Xenopus ovaries. Furthermore, abundant mRNAs such as histone transcripts (Fig. 2) and localized mRNAs, such as the oskar transcripts that occur at the posterior pole of Drosophila eggs, can be labeled with this method. Thus the user may try this protocol as a rapid first pass to examine mRNA localization before attempting more lengthy or costly techniques, such as nonfluorescent haptens (digoxigenin or biotin), chemical amplification methods such as tyramide signal amplification, or single molecule FISH.
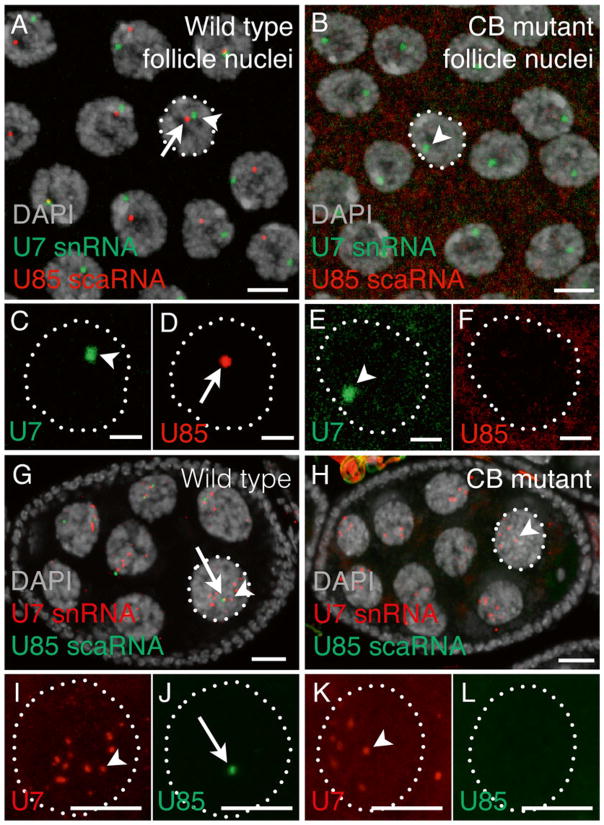
Fig. 1. FISH using U85 scaRNA (CB marker) and U7 snRNA (HLB marker) in wild-type and CB mutant Drosophila ovaries.
(a–f) Follicle (somatic cell) nuclei from stage 8 egg chambers labeled with an Alexa-488 U7 snRNA antisense probe (Alexa-488, green) and a Cy5 U85 scaRNA antisense probe (Cy5, pseudo-colored red). The arrows label CBs and the arrowheads label HLBs. Dotted white lines indicate nuclei enlarged in panels c–f. (a, c, d) y w flies are wild-type for CBs and HLBs. (b, e, f) Coilin199 /199 fl ies are coilin protein null mutants that lack CBs [4]. (g–l ) Stage 6 egg chambers showing germline nurse cell nuclei labeled with a Cy3 U7 snRNA antisense probe (Cy3, red ) and a Cy5 U85 scaRNA antisense probe (Cy5, pseudo-colored green ). The arrows label CBs and the arrowheads label HLBs. Dotted white lines indicate nuclei enlarged in panels i–l. (g, i, j ) Oregon-R fl ies are wild-type for CBs and HLBs. (h, k, l) WDR79MB10832/10832 fl ies are WDR79 protein-null mutants that lack CBs [5]. There are multiple HLBs in these nurse cells because the histone gene loci are dispersed due to loss of polyteny at this stage in development [6]. Cytoplasmic U7 snRNA bodies are U bodies [7]. Scale bar is 10 μm. Nuclear DNA was counterstained with DAPI (gray). Images are maximum intensity z-projections of multiple 0.5–1 μm optical sections obtained with a confocal microscope. Wild type and mutant specimens were treated identically and images were collected and processed with the same parameters
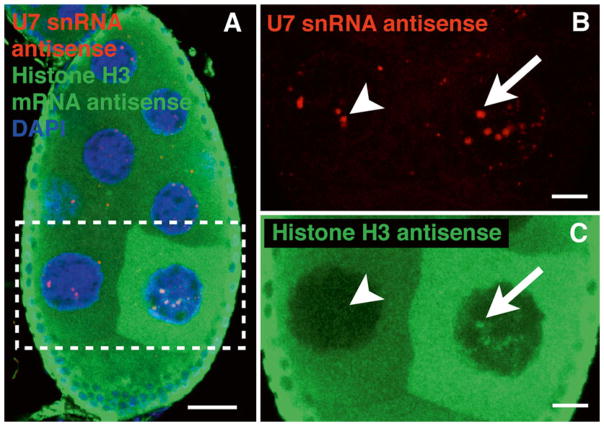
Fig. 2. FISH used to target histone H3 mRNA transcripts and U7 snRNA in a stage 7 wild-type and mutant Drosophila egg chamber.
(a) U7 snRNA antisense probe (Cy3, red) labels HLBs in all nurse cell nuclei, whereas histone H3 mRNA antisense probe (Cy5, green) labels one nurse cell more prominently than other nurse cells. Because histone transcription is replication dependent, these nurse cells accumulate histone transcripts at different times due to their asynchronous endocycles [8]. (b and c) Enlarged images of the nurse cells from the white box shown in panel (a). (b ) U7 snRNA antisense probe (red). (c ) Histone H3 mRNA anti-sense probe (green ). The histone H3 mRNA antisense probe is sensitive enough to detect cytoplasmic histone mRNA, as well as nascent transcripts at the histone locus in the transcribing nurse cell (arrow). In contrast, transcripts are not detected at HLBs in the other nurse cells (arrowhead), indicating that they are either not actively transcribing histone transcripts or the technique is not sensitive enough to detect them at this stage in the endocycle. Scale bar is 10 μm. Nuclear DNA was counterstained with DAPI (blue ). The image is a maximum intensity z-projection of multiple 0.5–1 μm optical sections obtained with a confocal microscope
2 Materials
2.1 Generating the Template DNA for In Vitro Transcription
Optional: pGEM T-easy vector (Promega).
Standard PCR reagents: usually included in a kit containing a DNA polymerase, dNTPs, and reaction buffers.
2.2 In Vitro Transcription Reaction
Nuclease-free or ultra pure water. Available commercially, or can be prepared chemically with DEPC as follows. DEPC- treated dH2O: Mix 2 ml of DEPC (diethyl pyrocarbonate) in 1 l of dH2O. Shake vigorously. Let sit in a hood for 1 h. Autoclave to inactivate the remaining DEPC.
QIAquick PCR purification kit (Qiagen).
20 mM CTP–ATP–GTP solution: Combine 4 μl each of 100 mM CTP, 100 mM ATP, and 100 mM GTP with 8 μl of dH2O.
10 mM UTP solution: Dilute the 100 mM UTP stock 1:10 in dH2O.
Fluorescent UTP or fluorescent CTP (see Note 1).
RNasin ribonuclease inhibitor.
T3, T7, or SP6 RNA polymerase.
5× transcription buffer (usually included with RNA polymerase).
DNAse I.
250 mM EDTA: Dissolve 9.3 g of Na2-EDTA·2H2O in approximately 80 ml of dH2O. Adjust the pH to 8.0 with 10 N NaOH (EDTA will not dissolve until pH is increased). Make up to 100 ml with dH2O. Autoclave.
GE Illustra MicroSpin G-50 Columns.
2.3 In Situ Hybridization of Whole-Mount Drosophila Ovaries
Grace’s Insect Medium (FBS-free).
Fine jeweler’s forceps.
Tungsten needles.
Dissecting microscope.
20 % paraformaldehyde (PFA): Weigh out 200 g PFA in a hood. Weigh out 0.5 g Na2CO3·H2O. Set a hot plate stirrer in the hood. Add Na2CO3 to ~800 ml dH2O. Stir. Then add 200 g PFA. Heat and stir to ~80 °C to get it into solution. Make up to 1 l with dH2O. Filter it through Whatman filter paper #1 in the hood since sometimes there is a lot of sludge. This stock is stable for many months at room temperature.
4 % PFA in Grace’s Insect Medium: Dilute 20 % PFA to 4 % in Grace’s Insect Medium.
5 % acetic acid (optional): Dilute v/v from glacial acetic acid.
20× phosphate buffered saline (PBS): 160 g NaCl, 4 g KCl, 12.2 g Na2HPO4 anhydrous, 4 g KH2PO4 in 1 l of dH2O.
9. 1× PBS: Dilute 1:20 with dH2O from a 20× stock.
In situ mix: Combine 25 ml of 50 % formamide, 12.5 ml of 20× SSC, 0.5 ml of 5 mg/ml heparin, 0.5 ml of 50 mg/ml yeast tRNA, 0.9 ml of 0.5 M citric acid, pH 6, 0.75 ml of 20 % Triton™ X-100, and 9.85 ml nuclease-free H2O to make a final volume of 50 ml.
Formamide.
20× SSC: Dissolve 175.3 g NaCl and 88.25 g sodium citrate (Na3C6H5O7·2H2O) in 1 l of dH2O, pH to 7.2 with 1 N HCl.
Heparin: Make a 5 mg/ml solution in dH2O.
Yeast tRNA (Roche, USA, Catalog # 10109517001): Make a 50 mg/ml solution in dH2O.
0.5 M citric acid, pH 6: Dissolve 14.8 g citric acid (C6H5O7Na3·2H2O) in 100 ml of dH2O (adjust to pH 6.0).
Triton™ X-100.
10 μg/ml DAPI (4′,6-diamidino-2-phenylindole dihydrochlo-ride): Dilute from 3.3 μg/μl stock in 70 % ethanol (stock can be stored in dark at 4 °C). The working stock (10 μg/ml) should be stored at room temperature in a dark bottle.
2.4 Mounting Labeled Tissue
Microscope glass slides and coverslips.
Phenylenediamine Stock Solution: Add 50 mg of phenylenedi-amine and 250 μl 20× PBS to 4.7 ml of dH2O. Vortex to dissolve. Bring to pH 9.0 with 1 N NaOH. Store at -70 °C.
Mounting Medium: Mix 5 ml of glycerol, 4 ml of dH2O and 1 ml of 10 mg/ml phenylenediamine Stock Solution. Aliquot rapidly to tubes on dry ice before storing at −70 °C.
Optional: Commercially available Mounting Media: ProLong Gold, Diamond antifade reagent, or VECTASHIELD.
Nail polish.
3 Methods
3.1 Generation of the Template DNA for In Vitro Transcription
Create an RNAse-free environment by using autoclaved tips and reagents, and nuclease-free or DEPC-treated dH2O in reactions. For suggestions in designing controls for this protocol, see Note 2. This protocol may be adapted to include immunostaining and FISH simultaneously or to target DNA loci instead of RNA as described below.
There are at least four options for obtaining the template DNA required to generate single-stranded RNA probes by in vitro transcription (see Note 3).
Obtain previously cloned plasmid DNA for use as standard markers, such as U7 snRNA for HLBs and U85 scaRNA for CBs [3] (see Note 4).
Clone the target genes for these standard markers into a vector containing T3, T7, or SP6 RNA polymerase promoters (see Note 5).
PCR amplify the target genes with oligos containing T3, T7, or SP6 RNA polymerase promoter sequences as overhangs (see Note 6).
Generate short RNA probes (15–65 nucleotides in length) from a synthetic DNA oligo that contains a T3, T7, or SP6 RNA polymerase promoter sequence at the 5′ end of a short template DNA sequence for the target gene (see Note 7).
3.2 In Vitro Transcription of the Directly Labeled Fluorescent RNA Probes
If using plasmid DNA as a template, linearize with the appropriate restriction enzyme and purify the DNA before proceeding with the in vitro transcription reaction (see Note 8).
Combine the reagents for the in vitro transcription reaction in a 1.7 ml centrifuge tube in the following order: 2 μl of 20 mM CTP–ATP–GTP, 1.5 μl of 10 mM UTP, 5 μl of 1 mM fluores-cent UTP, 1 μl of 40 mM RNAsin, 4 μl of 5× transcription buffer, 3.5 μl of nuclease-free H2O, 2 μl DNA (1 μg plasmid DNA or 100 ng PCR product), 1 μl of 50 U/μl enzyme (T3, T7, or SP6 polymerase) for a total volume of 20 μl. Assemble the reaction at room temperature, making sure to keep the RNA polymerase, NTPs, and RNase inhibitor on ice until use (see Note 9).
Mix well by tapping or vortex briefly. Pulse-spin. Incubate at 37 °C for 1.5–3 h.
Remove the template DNA by adding 1 μl of DNase I (dilute the stock solution 1:10 in DNase I buffer). Incubate at 37 °C for 10–20 min.
Inactivate the enzymes by adding 2 μl of 250 mM EDTA and incubating at 65 °C for 10–20 min.
Purify the RNA using a Sephadex column such as GE Illustra G50 spin columns, following manufacturer’s instructions (see Note 10).
Analyze the probes by UV spectrophotometry and gel electrophoresis. Test 1 μl of the in vitro transcribed RNA on a NanoDrop spectrophotometer to determine RNA concentration (the concentration should be approximately 300 ng/μl). Test 1 μl of the RNA on a denaturing agarose gel to verify the quality and length of the RNA (the RNA should run as a single band).
Store the probes at -20 °C (can be kept for many years). Working solutions of the 10× probe in the in situ mix can be kept at -20 °C for at least several months.
3.3 In Situ Hybridization of Whole-Mount Drosophila Ovaries
Dissect out Drosophila adult ovaries in a physiological saline solution such as Grace’s Insect Medium (see Notes 11 and 12 ).
Fix ovaries for 10 min in 4 % PFA in Grace’s Insect Medium at room temperature. While in the fixative, comb through the ovaries with fine forceps or tungsten needles to separate the ovarioles (see Notes 13 and 14).
Remove the PFA and wash 5 min at room temperature in 1× PBS (see Note 15).
Add 100 μl of 5 % Triton™ X-100 to the PBS before transferring egg chambers to a 500 μl microfuge tube using a cut off pipette tip (see Note 16).
Remove the PBS and wash the tissue for at least 10 min with 100 μl of in situ mix at room temperature.
Prepare the probe mix by diluting the probe (to empirically determined final concentration) and DAPI (to final concentration of 1 μg/ml) in the in situ mix (see Note 17). One can use as little as 10 μl of probe per sample in a microfuge tube. Remove the in situ mix and replace it with probe mix. Ensure that the tissue is completely submerged in probe and tap the tube well to mix.
Incubate the probe with the specimen at 42 °C for 6–16 h (see Note 18).
Remove the probe and wash for at least 10 min with 100 μl of in situ mix with 1 μg/ml DAPI at room temperature (see Note 19).
3.4 Mounting Labeled Tissue
Remove the in situ mix and replace it with 15 μl of Mounting Medium (see Note 20).
Pipette the tissue onto a microscope glass slide and distribute the ovarioles evenly with forceps under a dissecting microscope.
Add a 22 × 22 mm2 glass coverslip. To prevent egg chambers from being squashed, support the corners of the coverslip with a small amount of Vaseline or a Vaseline–paraffin wax mixture (melt equal weights of commercial Vaseline and par-affin wax, cool and store for use). Seal coverslip with commercial nail polish.
3.5 Combining Immunostaining and FISH
To perform the two methods simultaneously, modify the protocol in Subheading 3.3 as described below.
After fixation (step 4), proceed to a standard immunostaining protocol [3]. For a standard procedure, stain the tissue in primary and secondary antibodies overnight at room temperature.
Post-fix the tissue for 5 min at room temperature in 4 % PFA in 1× PBS.
Wash 2 × 5 min in 100 μl of 1× PBS at room temperature.
Continue with the FISH protocol starting at step 5 (blocking in in situ mix) (see Note 21).
3.6 To Hybridize to DNA Instead of RNA
Adapt the protocol in Subheading 3.3 to include steps to denature the target DNA as described below.
At step 6, denature the probe at 80 °C for 5 min and quench on ice until use.
At step 7 (hybridizing the probe to the specimen), denature the probe together with the specimen at 85 °C for 15 min before shifting to 42 °C for 6–16 h for hybridization (see Note 22).
Fig. 3. A schematic of the oligo design for incorporating RNA polymerase promoter sequences into template DNA for in vitro transcription.
The target genes sense strand is shown in black and the antisense strand in gray. The RNA polymerase promoters (T3, T7, or SP6) can be included as a 5′ overhang in the oligo design. To make an antisense probe, design the oligo to contain the reverse complement sequence (short gray line) downstream of the polymerase promoter (gray overhang line containing T3 RNA polymerase promoter sequence as an example). To make a sense probe, design the oligo to contain the coding sequence (short black line) downstream of the polymerase promoter (black overhang line containing T7 RNA polymerase promoter sequence as an example)
Acknowledgments
We thank Svetlana Deryusheva (Carnegie Institution for Science) for useful advice and enhancements to this protocol.
Footnotes
There are several choices for fluorescent UTP or CTP with different excitation/emissions (in parentheses). These include ChromaTide Alexa Fluor 488-5-UTP (490/520), Aminoallyl-UTP-ATTO-488 (501/523), Cy5-UTP (5 mM stock: use 1 μl/20 μl per reaction) (649/670), Cy3-CTP (550/570), and Alexa Fluor 546-14-UTP (555/570).
This FISH protocol allows the user to employ multiple useful and biologically meaningful controls. To test the specificity of the antisense probe, use a sense probe as a negative control (see Note 3) or hybridize the probe in a null mutant fly (for viable alleles) or inducible clonal mutant (for lethal alleles). For CB markers, use CB mutant flies such as the coilin or WDR79 protein-null flies [4, 5]. For the HLB marker U7 snRNA, use the U7 snRNA null mutant fly [9]. To control for proper identification of a nuclear body, label with two or more markers. If the markers are RNA targets, label FISH probes with fluorescent nucleotides with nonoverlapping spectra (such as Alexa 488 and Cy3). If labeling both a protein and an RNA target simultaneously, adapt the protocol as in Subheading 3.5.
In vitro transcribed single-stranded RNA probes are advantageous because one can generate strand-specific probes from the same cloned template DNA: one antisense probe that hybridizes to the target RNA and one sense probe that acts as a control for nonspecific hybridization. Additionally, single- stranded probes do not require extensive denaturing steps like double-stranded RNA or DNA probes. A possible disadvantage of RNA probes is that they are less stable than DNA probes, but in our experience, when properly stored at −20 °C, they can be kept for many years.
Our lab has standard CB and HLB markers cloned and ready for distribution upon request [3]. We typically use the U85 scaRNA as a CB marker and the U7 snRNA as an HLB marker. scaRNAs are the best markers for CBs because they are highly enriched in these bodies. CBs are not present in all cell types, and tend to be absent in undifferentiated cells such as stem cells or embryonic cells [2]. Spliceosomal U snRNAs and U7 snRNA are also present in cytoplasmic U bodies [7].
Because the RNA components of CBs and HLBs are generally short RNAs derived from intronless genes (such as scaRNAs and snRNAs), the target genes can be easily amplified by PCR from genomic DNA and cloned into a TA cloning vector such as Promega’s pGEM T-easy vector (contains T7 and SP6 phage RNA polymerase promoters). It is also straightforward to amplify the target gene by RT-PCR from total RNA. The short length of snRNAs and scaRNAs (<500 nt) makes them ideal for in situ hybridization probes because they readily penetrate whole tissues. If cloning into vectors that lack a T3, T7, or SP6 phage RNA polymerase promoter sequence, one can design oligos to include these sequences as a 5′ overhang (Fig. 3). The sequences to append the 5′ end of the oligos are: GCTAATACGACTCACTATAGGG for T7, GCAATTAACCCTCACTAAAGGG for T3, and GCATTTAGGTGACACTATAGA for SP6. The bold G in the above sequences represents the +1 nucleotide (start of transcription).
In vitro transcribed probes can also be made directly from a PCR product, provided that one includes an RNA polymerase promoter at the 5′ end of the oligo (Fig. 3). This is a quick and useful technique when screening for CB or HLB markers. One should sequence the PCR product to make sure that there are no other templates in the reaction, and it is best to clone the product into a vector for long-term storage.
Generating a short probe from a synthetic DNA oligo allows one to skip any PCR and cloning steps and works well for standard CB and HLB targets. The disadvantage to this method is the cost of synthesizing DNA oligos to include both the RNA polymerase promoter and the template sequence. Design the DNA oligo to contain 15–65 nt of target DNA template sequence downstream of a T3, T7, or SP6 RNA polymerase promoter (see Note 5 and Fig. 3). Modify the in vitro transcription reaction in Subheading 3.2, step 2 to include an anti-sense oligo against the T3, T7, or SP6 RNA polymerase phage promoter. By pairing these oligos together, one generates the double-stranded DNA template necessary for the phage RNA polymerase to function.
Template DNA can be purified by standard alcohol precipitation techniques, or can be rapidly purified using columns such as Qiagen’s QIAquick PCR purification kit (can also be used for restriction enzyme reactions).
The choice of a fluorescently conjugated nucleotide will depend on the lasers available for imaging the tissue. There are several fluorophores that we have found particularly bright and useful (see Note 1). One should select nonoverlapping emission spectra when performing 2-color or 3-color FISH. Note that some of the fluorophores are conjugated to CTP rather than UTP, so the in vitro reaction has to be adjusted accordingly (to modulate the ratio of the unlabeled to the labeled nucleotide).
A probe that has effectively incorporated the fluorescently conjugated nucleotide will have a hue corresponding to the color of the emission spectrum (for example, Alexa 488-UTP labeled probes will appear green).
The presence or absence of CBs and HLBs follows a stereotyped pattern during oogenesis [2]. However, the size, morphology, and number of non-membrane bound organelles can vary depending on the nutritional state of the animal [10, 11]. To ensure consistent analysis of nuclear bodies, it is best to dissect animals of similar age and genetic background raised on a standard food source. For example, collect 2 day-old flies and then supplement fly food with wet yeast for two additional days to stimulate ovary development.
The choice of solution used for dissection can affect the organization of the oocyte nucleus (germinal vesicle, GV). In salt solutions of low ionic concentration (below 10 mM), nuclear bodies are induced de novo, particularly in egg chambers that are damaged in the process of dissection [12]. Dissection in Grace’s Insect Medium preserves the morphology of nuclear bodies observed in live tissues. Furthermore, rapid dissection (less than 5 min, before fixation) is preferable for preserving morphology and preventing RNA degradation.
Any standard dissection and fixation technique will suffice. For example, dissect tissues in small 35 mm × 10 mm petri dishes or well slides, and transfer them with forceps to a separate well or dish that contains 100 μl of fixative, prepared fresh from a 20 % PFA stock. Drosophila ovaries are sufficiently fixed in as little as 10 min. Longer fixation periods are not necessary to preserve ovary morphology, and have been generally avoided because of the possibility of reduced permeability of the tissue to probe.
For better preservation of RNA, add acetic acid to the fixative to a final concentration of 0.5–2.5 %. This improves the brightness of the FISH signal but leaves the tissue more brittle and prone to damage. Acetic acid is not essential when the RNA target is abundant or densely localized, as it is in CBs or HLBs. Moreover, fixation with acetic acid is generally not compatible with immunostaining.
If dissecting in small petri dishes, use enough 1× PBS to fill the dish (approximately 3 ml).
Drosophila tissues stick to plastic surfaces unless solutions contain a detergent. To achieve a more uniform penetration of RNA probes into the tissues when examining younger egg chambers (up to stage 10), remove the more mature chambers at this point (stages 11–14).
Probes generated by in vitro transcription as described in Subheading 3.2 are generally at a concentration of 300 ng/μl. Probes against RNA targets that are concentrated in nuclear bodies such as CBs and HLBs can be diluted as much as 1000× for FISH. Probes against diffusely localized RNA or lower abundance RNA targets should be diluted only 20–200×. DAPI is added to the probe mix to provide a robust fluores-cent counterstain for nuclear DNA. If using minimal volumes of probe mix, such as 10 μl/sample, remove all wash solution from the specimen before adding the probe.
An incubation temperature of 42 °C is appropriate for hybridization of RNA probes (in 50 % formamide) to RNA targets in situ. Individual probes can be tested for optimal hybridization in the range of 37–52 °C (the higher the temperature, the more stringent the hybridization conditions). One can complete the procedure in 1 day (6 h incubation) or opt for an overnight incubation (up to 16 h) to analyze the next day. Overnight incubations are preferred for lower abundance targets, but are not necessary for most CB or HLB markers. For hybridization to RNA targets, incubations longer than 16 h may diminish the signal intensity due to degradation of the RNA targets. When using shorter RNA probes (such as 30–80 nucleotides in length), the hybridization incubation time can be reduced to 30 min–2 h because shorter probes penetrate the tissue better.
This short wash step is sufficient for examining CB and HLB markers because the RNA targets are highly localized. If experiencing high background, begin troubleshooting by decreasing the concentration of the probe before increasing the number, duration and/or temperature of washes.
We mount specimens in a phenylenediamine solution, which is cost effective but turns brown in a few weeks, even at -20 °C. For permanent preparations use commercially available mounting media such as VECTASHIELD, ProLong Gold, or ProLong Diamond.
The FISH signal may be diminished due to degradation of target RNA during immunostaining steps. If so, one can adapt the FISH protocol by using the probe at a higher concentration, or conducting the antibody staining steps at 4 °C.
The temperature and duration for the denaturation step affects the morphology of the tissue. Other denaturation treatments that work include 80 °C for 10 min or 90 °C for 5 min. For hybridization to DNA targets, the incubation time can be extended to several days because the DNA targets are more stable than RNA targets, and the RNA probes are stable under these conditions. Both sense and antisense probes will hybridize to target DNA; hybridizing to target DNA is a useful method to test the specificity of the sense probe. These directly labeled fluorescent probes are sensitive enough to robustly label large gene clusters such as the histone genes. Other gene loci have not yet been tested.
References
- 1.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nizami ZF, Deryusheva S, Gall JG. Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harb Symp Quant Biol. 2010;75:313–320. doi: 10.1101/sqb.2010.75.005. [DOI] [PubMed] [Google Scholar]
- 3.Liu J-L, Murphy C, Buszczak M, et al. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J-L, Wu Z, Nizami Z, et al. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20:1661–1670. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deryusheva S, Gall JG. Novel small Cajal-body-specific RNAs identified in Drosophila: probing guide RNA function. RNA. 2013;19:1802–1814. doi: 10.1261/rna.042028.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126:293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- 7.Liu JL, Gall JG. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc Natl Acad Sci USA. 2007;104:11655–11659. doi: 10.1073/pnas.0704977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White AE, Leslie ME, Calvi BR, et al. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godfrey AC, Kupsco JM, Burch BD, et al. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckingham M, Liu JL. U bodies respond to nutrient stress in Drosophila. Exp Cell Res. 2011;317:2835–2844. doi: 10.1016/j.yexcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Shimada Y, Burn KM, Niwa R, et al. Reversible response of protein localization and microtubule organization to nutrient stress during Drosophila early oogenesis. Dev Biol. 2011;355:250–262. doi: 10.1016/j.ydbio.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer AB, Gall JG. An inducible nuclear body in the Drosophila germinal vesicle. Nucleus. 2011;2:403–409. doi: 10.4161/nucl.2.5.17250. [DOI] [PMC free article] [PubMed] [Google Scholar]