Here, Elbarbary et al. provide new insights into Tudor-SN (TSN)-mediated miRNA decay (TumiD), in which the nuclease TSN promotes the decay of miRNAs that contain CA and/or UA dinucleotides. They show that cellular TumiD requires RNA helicase UPF1 and, by using miR-seq in T24 bladder cancer cells, that the degradation of ∼50% of candidate TumiD targets in T24 cells is augmented by UPF1.
Keywords: mass spectrometry, miRNA–mRNA duplexes, RISC constituents, RNP remodeling, miR-seq, bladder cancer, invasiveness
Abstract
While microRNAs (miRNAs) regulate the vast majority of protein-encoding transcripts, little is known about how miRNAs themselves are degraded. We recently described Tudor-staphylococcal/micrococcal-like nuclease (TSN)-mediated miRNA decay (TumiD) as a cellular pathway in which the nuclease TSN promotes the decay of miRNAs that contain CA and/or UA dinucleotides. While TSN-mediated degradation of either protein-free or AGO2-loaded miRNAs does not require the ATP-dependent RNA helicase UPF1 in vitro, we report here that cellular TumiD requires UPF1. Results from experiments using AGO2-loaded miRNAs in duplex with target mRNAs indicate that UPF1 can dissociate miRNAs from their mRNA targets, making the miRNAs susceptible to TumiD. miR-seq (deep sequencing of miRNAs) data reveal that the degradation of ∼50% of candidate TumiD targets in T24 human urinary bladder cancer cells is augmented by UPF1. We illustrate the physiological relevance by demonstrating that UPF1-augmented TumiD promotes the invasion of T24 cells in part by degrading anti-invasive miRNAs so as to up-regulate the expression of proinvasive proteins.
MicroRNAs (miRNAs), which are small and noncoding, constitute an intricate network of gene regulatory events that elicit the degradation and/or translational repression of mRNAs (Gurtan and Sharp 2013; Iwakawa and Tomari 2015). Despite there being good understanding of miRNA biogenesis and mechanisms of action (Ha and Kim 2014), there is a relative dearth of information on how miRNAs are degraded.
Human TSN, an evolutionarily conserved nuclease that contains a Tudor domain and five staphylococcal/micrococcal-like nuclease (SN) domains, was the first constituent of the RNA-induced silencing complex (RISC) shown to have endonuclease activity (Caudy et al. 2003). While TSN was initially proposed to be the endonuclease that cleaves miRNA targeted mRNAs, later reports demonstrated that AGO2 manifests this cleavage activity (Hammond 2005). We demonstrated recently that TSN degrades functional miRNAs in a pathway that we named Tudor-mediated miRNA decay (TumiD) (Elbarbary et al. 2017). In cells, TumiD does not depend on biogenesis steps that precede miRNA:miRNA* (i.e., miR-5p:miR-3p) duplex formation. Consistent with this, cellular TumiD effectively degrades exogenously introduced miRNA mimics of bona fide TumiD targets; i.e., miRNAs that contain at least one CA or UA dinucleotide, preferably residing between their 5′-most and 3′-most 5 nucleotides (nt) (Elbarbary et al. 2017). As evidence for the evolutionary conservation of TumiD, all mouse miRNAs identified as orthologs of human TumiD targets characterized in human embryonic kidney (HEK) 293T cells have CA and/or UA dinucleotides at identical positions and thus are predicted to be TumiD targets (Elbarbary et al. 2017). Prior to our work, TSN was known to facilitate the G1-to-S phase transition in mouse embryonic fibroblasts (Su et al. 2015). We showed that TSN does likewise in HEK293T cells by degrading miRNAs that down-regulate the expression of proteins critical for the G1-to-S-phase transition (Elbarbary et al. 2017). Consistent with this, TSN is overexpressed in various types of cancers, promoting carcinogenesis by diverse transcriptional and post-transcriptional mechanisms (Jariwala et al. 2015).
RNA helicases often use ATP to unwind dsRNA structures and function in RNA metabolism through diverse mechanisms that rely on their ability to remodel the composition of RNA–protein complexes (RNPs) (Bourgeois et al. 2016). Several helicases play roles in RNAi through variable yet incompletely understood mechanisms, some independently of their helicase activities (Ambrus and Frolov 2009).
The RNA helicase UPF1 is the central factor in nonsense-mediated mRNA decay (NMD) and Staufen-mediated mRNA decay (SMD) (Park and Maquat 2013; Karousis et al. 2016; Kurosaki and Maquat 2016; Ottens and Gehring 2016; Popp and Maquat 2016). While UPF1 cycles on and off cellular RNAs that are accessible to binding in a mechanism that depends on ATP hydrolysis (Kurosaki et al. 2014; Lee et al. 2015; Durand et al. 2016) and likely involves movement along RNAs in a 5′-to-3′ direction (Franks et al. 2010; Fiorini et al. 2015), regulated UPF1 binding to an NMD target promotes mRNA decay by remodeling the mRNA and its associated proteins; i.e., by remodeling mRNP (Franks et al. 2010; Gregersen et al. 2014).
Here we report that UPF1 coimmunoprecipitates with TSN in HEK293T cell lysates and plays a role in cellular TumiD. Consistent with this, TSN and UPF1 coimmunoprecipitate with RISC constituents AGO2 and the trinucleotide repeat-containing protein TNRC6A, also called GW182. Knocking down UPF1 up-regulates the levels of miRNAs degraded by TumiD to almost the same extent as knocking down TSN. Additionally, miRNAs degraded by TumiD are detectable in tandem immunoprecipitations of TSN and UPF1. UPF1 function in cellular TumiD depends on its helicase activity. While in vitro synthesized single-stranded miRNAs are susceptible to recombinant TSN-mediated degradation in the absence of UPF1, efficient degradation of their cellular counterparts requires UPF1, supporting the idea that UPF1 remodels miRNPs during TumiD. Adding UPF1 to miRNA–mRNA duplexes dissociates miRNAs from their mRNA targets, rendering them susceptible to TSN cleavage. UPF1 retains its ability to unwind miRNA–mRNA duplexes even when the miRNAs are bound by AGO2, suggesting that UPF1 promotes TumiD by dissociating RISC-bound miRNAs from their mRNA targets.
We used deep sequencing of miRNAs (miR-seq) to globally define candidate TumiD targets in human T24 bladder cancer cells and show that ∼50% of miRNAs that are up-regulated by TSN knockdown are also up-regulated by UPF1 knockdown. Illustrating biological activity, we demonstrate that TSN and UPF1 enhance T24 bladder cancer cell invasion by mediating the degradation of a cohort of anti-metastatic miRNAs that down-regulate the expression of proinvasion proteins.
Results
TSN and UPF1 form a complex in human cells
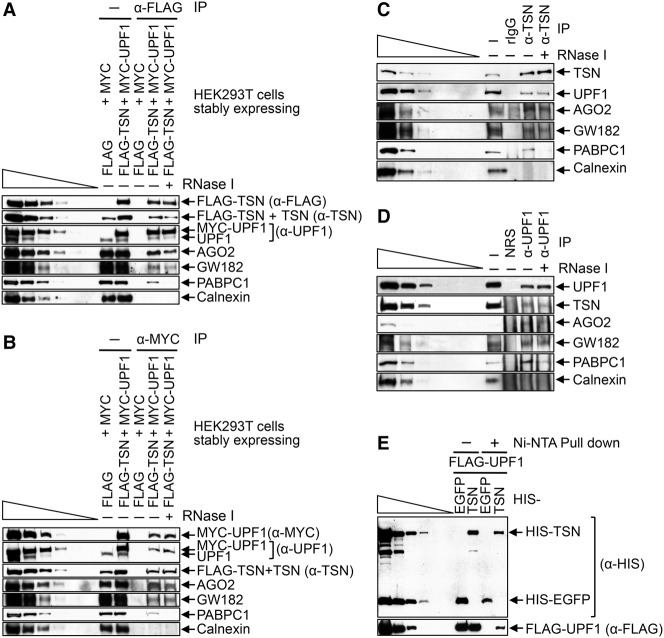
While examining the mechanism of TumiD, we found that cellular UPF1 coimmunoprecipitates with cellular TSN in HEK293T cell lysates (data not shown). To investigate this interaction, we generated HEK293T cells stably expressing Flag-TSN and MYC-UPF1, each at a level approximating the level of its cellular counterpart (Fig. 1A,B), and performed mass spectrometry on the proteins obtained after anti-Flag immunoprecipitation and, separately, anti-MYC immunoprecipitation in both the presence and absence of RNase I. We chose to immunoprecipitate tagged proteins, since immunoprecipitation efficiencies are far superior to those obtained using an antibody to either TSN or UPF1 (data not shown). The results revealed that UPF1 was among the top five proteins that coimmunoprecipitated with Flag-TSN (Supplemental Table S1), and TSN was among the top five proteins coimmunoprecipitated with MYC-UPF1 (Supplemental Table S2). Western blotting confirmed these coimmunoprecipitations (co-IPs) and also the co-IP of RISC constituents AGO2 and GW182 (Fig. 1A,B), in agreement with previous reports (Caudy et al. 2003; Jin et al. 2009). These data provide the first indication that UPF1 may function in TumiD.
Figure 1.
Evidence that TSN and UPF1 form a complex in HEK293T cells. (A) Western blotting of formaldehyde cross-linked HEK293T cells that stably express both Flag-TSN and MYC-UPF1 or, as a negative control, Flag and MYC. Western blotting was performed before (−) or after immunoprecipitation using anti-Flag in the absence (−) or presence (+) of RNase I. Here and elsewhere, lanes below the wedge analyze threefold dilutions of lysate. (B) As in A except that immunoprecipitations were performed using anti-MYC. (C) As in A except that lysates of untransfected HEK293T cells were used, and immunoprecipitations were performed using anti-TSN or, as a negative control, rabbit IgG (rIgG). (D) As in C except that immunoprecipitations were performed using anti-UPF1 or, as a negative control, nonimmunized rabbit serum (NRS). (E) HIS pull-downs of Escherichia coli-produced HIS-TSN or HIS-EGFP (HIS-tagged enhanced green fluorescence protein) in the presence of baculovirus-produced Flag-UPF1. Results are representative of three or more independently performed experiments.
We corroborated the finding that cellular TSN and UPF1 coimmunoprecipitate in two ways. First, Western blotting of immunoprecipitations of cellular TSN revealed the co-IP of cellular UPF1 as well as AGO2 and GW182, each in an RNase I-resistant manner (Fig. 1A–C). Notably, the co-IP of poly(A)-binding protein C1 (PABPC1)was largely RNase I-sensitive as expected, and the failure to detect calnexin in any immunoprecipitation confirmed immunoprecipitation specificity (Fig. 1C). Second, immunoprecipitations of cellular UPF1 revealed the co-IP of cellular TSN, AGO2, and GW182 in the presence of RNase I (Fig. 1D). Importantly, TSN and UPF1 likely interact directly in human cells, considering that baculovirus-produced and purified Flag-UPF1 was pulled down with Escherichia coli-produced and purified HIS-TSN but not with the negative control protein HIS-EGFP (HIS-tagged enhanced green fluorescence protein) (Fig. 1E).
UPF1 promotes the TumiD of mature miRNAs
Given that TSN and UPF1 form a complex in human cells and coimmunoprecipitate with RISC components (Fig. 1) and considering the role of TSN in TumiD (Elbarbary et al. 2017), it was possible that UPF1 also functions in TumiD. Alternatively or additionally, UPF1 might regulate pri-miRNA and/or pre-miRNA processing given the precedent that RNA helicases can influence the rate of miRNA processing (e.g., see Davis et al. 2008; Michlewski and Cáceres 2010). To test these possibilities, we measured the levels of individual TumiD targets and their pri-miRNAs and pre-miRNAs in HEK293T cells transiently transfected with control siRNA, UPF1 siRNA, or, as a positive control, TSN siRNA.
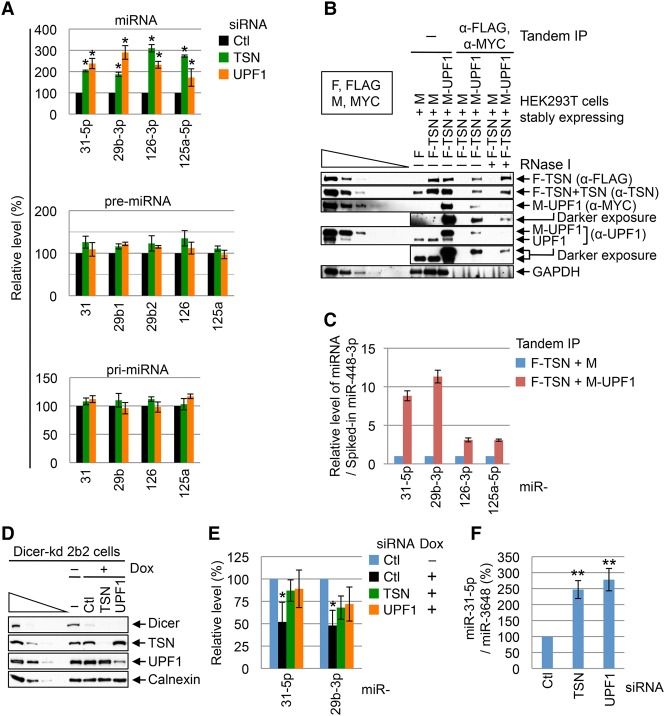
Like TSN knockdown, UPF1 knockdown in HEK293T cells (Supplemental Fig. S1A) increased the abundance of miR-31-5p, miR-29b-3p, miR-126-3p, and miR-125a-5p, which are bona fide TumiD targets (Elbarbary et al. 2017), approximately twofold to threefold without affecting the level of the corresponding pri-miRNA or pre-miRNA (Fig. 2A). The observed effects of UPF1 are not attributable to its role in NMD or SMD, since down-regulating the UPF2 or UPF3X NMD factor or the Staufen1 SMD factor did not up-regulate the level of the studied miRNAs (data not shown).
Figure 2.
UPF1 promotes TumiD in HEK293T cells. (A) Histogram of RT-qPCR quantitations of miRNAs and pre-miRNAs (using fractions enriched in <200-nt RNAs) and pri-mRNAs (using fractions enriched in >200-nt RNAs) from HEK293T cells transfected with control (Ctl) siRNA, TSN siRNA, or UPF1 siRNA. The level of each miRNA or pre-miRNA was normalized to the level of U6 snRNA, the level of each pri-miRNA was normalized to the level of β-actin mRNA, and normalized levels in control siRNA-treated cells are defined as 100. Normalizations to the level of U61 snoRNA gave results comparable with those obtained after normalizing to U6 snRNA (data not shown). (B) Western blotting of tandem immunoprecipitations described in Supplemental Figure S1B. “F” denotes Flag, and “M” denotes MYC. (C) Histogram of RT-qPCR quantitations of miRNAs extracted from tandem immunoprecipitations analyzed in B, where the level of each miRNA was normalized to the level of spiked-in miR-448-3p, and the normalized level in the F-TSN + M immunoprecipitation is defined as 1. (D) Western blotting of lysates of Dicer knockdown HEK293 2b2 cells that were harvested 96 h after the addition of doxycycline (Dox), which induces Dicer shRNA production. (E) Histogram of RT-qPCR quantitations of miRNAs derived from cells analyzed in D. The level of each miRNA was normalized to the level of U6 snRNA, and the normalized level in cells not treated with Dox is defined as 100. (F) Histogram of RT-qPCR quantitations of miRNAs derived from HEK293T cells transfected with the designated siRNA together with 1 nM miR-31-5p mimic and 1 nM miR-3648 mimic. The cellular uptake of each mimic was halted 4 h after transfection to allow assays of miRNA turnover. The level of miR-31-5p was normalized to the level of miR-3648, which is not a TumiD target (Elbarbary et al. 2017). The normalized level in control siRNA-treated cells is defined as 100. All results are representative of three or more independently performed experiments. (*) P < 0.05; (**) P < 0.01. n ≥ 3. Error bars represent ±S.D.
To investigate whether the interaction of cellular TSN and UPF1 occurs on miRNPs, we performed a tandem immunoprecipitation (anti-Flag followed by anti-MYC) (Supplemental Fig. S1B) using lysates of HEK293T cells stably expressing Flag-TSN and MYC-UPF1 (Fig. 2B). The results revealed that all bona fide TumiD targets assayed were enriched in the tandem immunoprecipitation (Fig. 2C), indicating that TSN and UPF1 cobind miRNPs.
The simplest interpretation of our results is that UPF1 promotes TumiD per se. We knew that UPF1 is not required for TumiD in vitro, since recombinant TSN alone is sufficient to degrade naked as well as AGO2-loaded in vitro synthesized miRNAs (Elbarbary et al. 2017). However, to further substantiate that UPF1 promotes TumiD in cells, we used HEK293T cells in which Dicer could be conditionally knocked down, blocking miRNA biogenesis (Fig. 2D) and allowing us to examine miRNA degradation without confounding effects from synthesis rates. Results indicated that upon Dicer knockdown, miR-31-5p and miR-29b-3p accumulated in UPF1 knockdown or TSN knockdown cells relative to cells transfected with control siRNA (Fig. 2E). Consistent with this, down-regulating either TSN or UPF1 promoted the accumulation of the miR-31-5p TumiD target that was exogenously introduced into cells as a miR-31-5p:miR-31-3p duplex (Fig. 2F). We conclude that TSN and UPF1 coordinately mediate cellular TumiD in a mechanism that can be uncoupled from the pri-miRNA and pre-miRNA processing steps that precede miRNA:miRNA* duplex formation
UPF1 helicase activity is required for efficient cellular TumiD
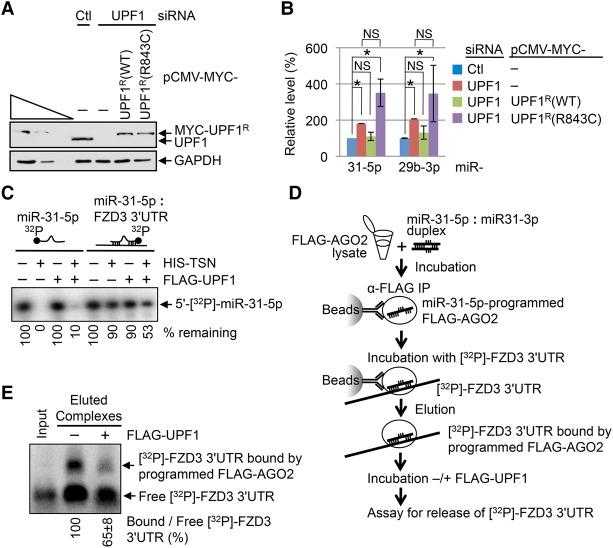
To determine whether the helicase activity of UPF1 functions in TumiD, we assayed HEK293T cells cotransfected with control siRNA or UPF1 siRNA and a plasmid producing either MYC-UPF1R wild type or MYC-UPF1R(R843C) (Fig. 3A), the latter of which lacks helicase activity (Supplemental Fig. S2A,B; Kurosaki et al. 2014) because it fails to efficiently bind ATP (Kurosaki et al. 2014). Assays of endogenous TumiD targets demonstrated that MYC-UPF1R, but not MYC-UPF1R(R843C), rescued endogenous UPF1 function in TumiD (Fig. 3B). MYC-UPF1R(G495R/G497E), another helicase-dead UPF1 variant (Kurosaki et al. 2014), also failed to rescue endogenous UPF1 function in TumiD, whereas MYC-UPF1R(4SA), a UPF1 variant that is hypophosphorylated but maintains wild-type helicase activity (Kurosaki et al. 2014), did rescue function (data not shown). We conclude that efficient cellular TumiD requires UPF1 helicase activity.
Figure 3.
UPF1 helicase activity is essential for efficient cellular TumiD. (A) Western blotting of total cell lysates of HEK293T cells transiently transfected with control (Ctl) or UPF1 siRNA and the specified plasmid. Superscript R denotes siRNA resistance. (B) Histogram of RT-qPCR quantitations of miRNAs derived from samples analyzed in A. The level of each miRNA was normalized to the level of U6 snRNA, and the normalized level in control siRNA-treated and pCMV-MYC-treated cells is defined as 100. (C) 5′-[32P]-labeled miR-31-5p, either single-stranded or in duplex with in vitro synthesized 80 nt of the frizzled class receptor 3 (FZD3) mRNA 3′ untranslated region (UTR), was incubated with HIS-TSN, Flag-UPF1, or a mixture of HIS-TSN and Flag-UPF1. (D) Flow chart depicting experiments where lysates of HEK293T cells stably expressing 3xFlag-AGO2 were incubated with the miR-31-5p:miR-31-3p duplex, from which 3xFlag-AGO2 was immunoprecipitated on anti-Flag-bound beads and subsequently incubated with internally labeled [32P]-FZD3 3′ UTR. 3xFlag-AGO2-bound miR-31-5p:FZD3 3′ UTR complexes were eluted from beads using Flag peptide, subsequently incubated with or without Flag-UPF1, and electrophoresed in a native agarose gel. (E) Results derived from experiments depicted in D. The ratio of FZD3 3′ UTR base-paired with programmed 3xFlag-AGO2 (i.e., bound FZD3 3′ UTR) to free FZD3 3′ UTR in the absence of Flag-UPF1 is defined as 100. Input represents free [32P]-FZD3 3′ UTR used to form complexes. As expected, bound [32P]-FZD3 3′ UTR was undetectable unless the lysates were incubated with the miR-31-5p:miR-31-3p duplex (data not shown). All results are representative of three or more independently performed experiments. (*) P < 0.05; (NS) statistically nonsignificant (P > 0.05). n ≥ 3. Error bars represent ±S.D.
UPF1 helicase activity may promote cellular TumiD by reorganizing RISC, which includes dissociating miRNAs from their target mRNAs to increase miRNA accessibility to TSN-mediated decay. To test these hypotheses, we first assayed TSN for the ability to degrade dsRNAs. We generated in vitro synthesized 5′-[32P]-miR-31-5p duplexed to its mRNA-binding site; i.e., duplexed to 80 nt of the 3′ untranslated region (UTR) of frizzled class receptor 3 (FZD3) mRNA (Fig. 3C). Compared with 5′-[32P]-miR-31-5p alone, the 5′-[32P]-miR-31-5p:FZD3 3′ UTR duplex was resistant to degradation by HIS-TSN (Fig. 3C), in agreement with previous demonstrations that inosine-lacking dsRNAs are resistant to TSN-mediated degradation (Scadden 2005). However, adding baculovirus-produced Flag-UPF1 (Supplemental Fig. S2C), whose helicase activity is capable of unwinding dsRNA with a 5′ overhang (Park et al. 2013), released 5′-[32P]-miR-31-5p from the FZD3 3′ UTR (Supplemental Fig. S2D) and also increased the susceptibility of 5′-[32P]-miR-31-5p to HIS-TSN-mediated decay (Fig. 3C).
To test whether UPF1 maintains its ability to release a miRNA from its target mRNA when the miRNA is bound by AGO2, we incubated 3xFlag-AGO2-bound miR-31-5p duplexed to internally [32P]-labeled FZD3 3′ UTR (Fig. 3D) with or without Flag-UPF1 and found that Flag-UPF1 released ∼35% of FZD3 3′ UTR from 3xFlag-AGO2-bound miR-31-5p (Fig. 3E). We conclude that cellular UPF1 promotes TumiD at least in part by releasing AGO2-bound miRNAs from their mRNA targets.
miR-seq of T24 bladder cancer cells
Both TSN and UPF1 play roles in tumorigenesis (Jariwala et al. 2015; Lindeboom et al. 2016), which spurred us to investigate whether these roles are mediated through TumiD. To this end, we knocked down TSN and, separately, UPF1 in human T24 bladder cancer cells and used miR-seq to globally identify miRNAs regulated by each protein. Pair-wise correlation among normalized reads generated by five biological miR-seq replicates gave a Pearson correlation coefficient of ≥0.95 (Supplemental Fig. S3A), indicating high correlation among replicates. Knocking down either TSN or UPF1 resulted in significant up-regulation of 131 miRNAs (adjusted P-value of <0.05), 65 of which were in common in both pools of up-regulated miRNAs (referred to here as the common pool) (Fig. 4A,B; Supplemental Table 3). Therefore, ∼50% of candidate TumiD targets in T24 cells are coregulated by TSN and UPF1, providing further support for our model that UPF1 promotes TumiD.
Figure 4.
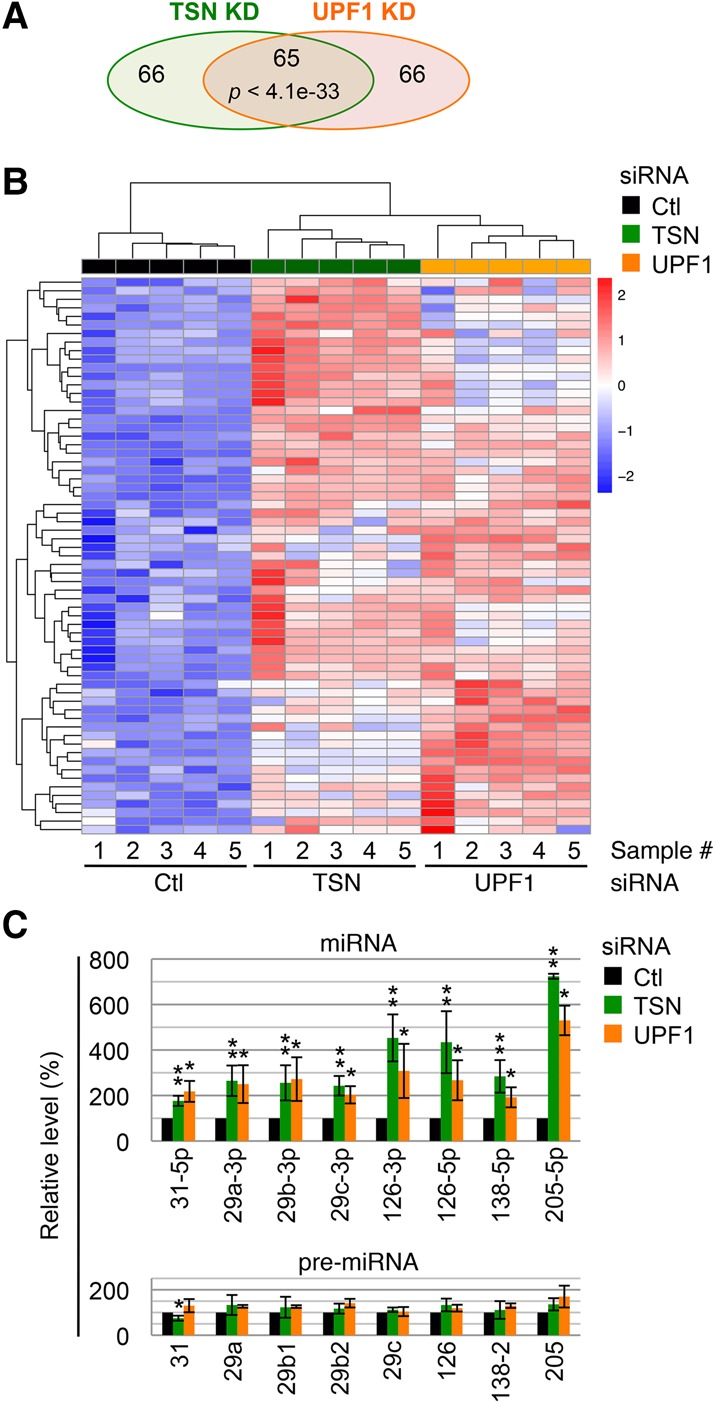
UPF1 coregulates ∼50% of candidate TumiD targets in T24 cells. (A) A Venn diagram representation of miR-seq results showing the numbers of miRNAs significantly up-regulated (P-value < 0.05) in TSN knockdown or UPF1 knockdown cells relative to control (Ctl) siRNA transfected cells. The significance of the overlap between up-regulated miRNAs from either knockdown was calculated using the hypergeometric test and a total of 1102 expressed miRNAs. (B) Heat map of hierarchically clustered miRNA expression in the miR-seq data showing the common pool. The color key represents row-scaled rLog transformed expression values. (C) Histogram of RT-qPCR quantitations of miRNAs and pre-miRNAs. Quantitations were performed using fractions enriched in <200-nt RNAs extracted from T24 cells transfected with control siRNA, TSN siRNA, or UPF1 siRNA. The level of each miRNA or pre-miRNA was normalized to the level of U6 snRNA, and normalized levels in control siRNA-treated cells are defined as 100. All results are representative of three or more independently performed experiments. (*) P < 0.05; (**) P < 0.01. n ≥ 3. Error bars represent ±S.D.
Using RT-qPCR, we analyzed 11 miRNAs in the common pool, eight of which proved to be bona fide TumiD targets, as evidenced by their significant up-regulation without concomitant changes in the levels of their corresponding pre-miRNAs (Fig. 4C). The remaining three miRNAs showed different results: miR-137 was up-regulated at both its mature and pre-miRNA levels (Supplemental Fig. S3B), which obscured our ability to conclude that it is a TumiD target; miR-15a-5p was up-regulated to a level comparable with that measured by miR-seq but without statistical significance (Supplemental Fig. S3C); and miR-32-5p was not detectably up-regulated (Supplemental Fig.S3C). All bona fide TumiD targets were also significantly up-regulated upon UPF1 knockdown without concomitant significant changes in the levels of their corresponding pre-miRNAs (Fig. 4C).
The common pool showed significant enrichment for miRNAs involved in carcinoma when analyzed using miRNA enrichment analysis and annotation (Supplemental Table S4; Backes et al. 2016). Moreover, analyzing experimentally validated targets of the six miRNAs whose levels were up-regulated threefold or more in either TSN knockdown or UPF1 knockdown cells using Diana Tools mirPath version 3 (Vlachos et al. 2012) revealed enrichment for genes involved in cancer pathways (Supplemental Table S5). These findings encouraged us to study the role of TumiD in carcinogenesis.
UPF1 promotes TumiD of biologically functional anti-invasive miRNAs in bladder cancer cells
To begin to investigate TumiD in cancer, we noted that miR-31-5p and miR-29b-3p are well-studied anti-invasive miRNAs in breast cancer cells (Augoff et al. 2011; Chou et al. 2013), and miR-126-3p has been shown to inhibit bladder cancer cell invasion (Jia et al. 2014). Since all three miRNAs are bona fide TumiD targets whose turnover is promoted by TSN and UPF1 in T24 cells, as indicated by both miR-seq and RT-qPCR (Fig. 4; Supplemental Table S3), we examined the functional significance of their regulation by TSN and UPF1 in T24 cells.
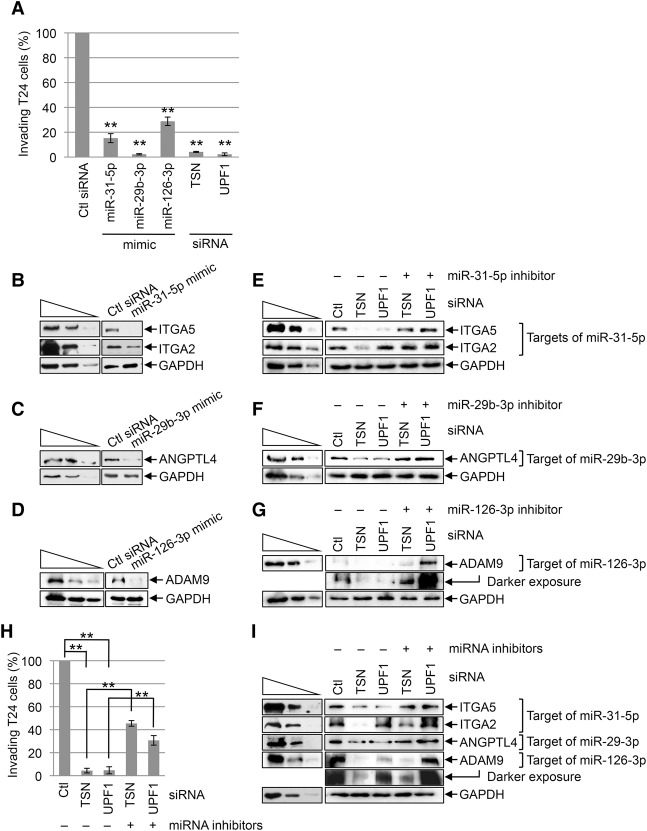
We first tested for the anti-invasive function of the three miRNAs using T24 cells that had been transfected with control siRNA, TSN siRNA, UPF1 siRNA, or a mimic of each miRNA. Relative to control siRNA, T24 cell invasion was inhibited ∼75%–95% by each miRNA mimic or siRNA (Fig. 5A; Supplemental Fig. S4A). Since we limited our analysis to 3–4 d after transfection, the observed decrease in T24 cell invasion upon transient knockdown of TSN or UPF1 was not due to changes in the rate of cell proliferation (Supplemental Fig. S4B).
Figure 5.
TumiD promotes bladder cancer cell invasion. (A) Histogram representing quantitation of T24 cell invasion (Supplemental Fig. S4A). Cells were transfected with the designated siRNA or miRNA mimic, and the number of invading control (Ctl) siRNA transfected cells is defined as 100. (B) Western blotting of lysates of T24 cells transfected with control siRNA or a miR-31-5p mimic. (C) As in B except cells were transfected with control siRNA or a miR-29b-3p mimic. (D) As in B except cells were transfected with control siRNA or a miR-126-3p mimic. (E) Western blotting of T24 cells transfected with the designated siRNA along with a control inhibitor (−) or miR-31-5p inhibitor. (F) As in E except cells were transfected with a control or miR-29b-3p inhibitor. (G) As in E except cells were transfected with a control or miR-126-3p inhibitor. (H) As in A except cells were transfected with the designated siRNA along with either a control inhibitor (designated “−” miRNA inhibitors) or a cocktail of miR-31-5p, miR-29b-3p, and miR-126-3p inhibitors (designated “+” miRNA inhibitors) (see also Supplemental Fig. S4C). (I) As in E except cells were transfected with a control inhibitor (−) or a cocktail of miR-31-5p, miR-29b-3p, and miR-126-3p inhibitors (+ miRNA inhibitors). All results are representative of three or more independently performed experiments. (**) P < 0.01. n ≥ 3. Error bars represent ±S.D.
To gain insight into mechanism, we found that the miR-31-5p mimic down-regulated the levels of integrin subunit α5 (ITGA5) and ITGA2 proteins (Fig. 5B), the miR-29b-3p mimic down-regulated the level of angiopoietin like 4 (ANGPTL4) protein (Fig. 5C), and the miR-126-3p mimic down-regulated the level of ADAM metallopeptidase domain 9 (ADAM9) protein (Fig. 5D), all of which are proinvasive proteins (Augoff et al. 2011; Chou et al. 2013; Wang et al. 2013). The up-regulation of miR-31-5p, miR-29b-3p, and miR-126-3p levels observed in T24 cells after TSN knockdown or UPF1 knockdown (Fig. 4C; Supplemental Table 3) was accompanied by down-regulation of the four prometastatic proteins (Fig. 5E–G) with the exception of the ITGA2 protein, which was down-regulated after TSN knockdown but up-regulated after UPF1 knockdown, possibly because ITGA2 mRNA consists of splicing variants that undergo NMD (data not shown). Transfecting T24 cells with a miR-31-5p, miR-29b-3p, or miR-126-3p inhibitor rescued expression of the corresponding prometastatic proteins (Fig. 5E–G), indicating that TSN and UPF1 regulate the levels of these proteins via miRNAs. Additionally, while TSN or UPF1 knockdown inhibited T24 cell invasion to ∼5%–10% of control knockdown, transfecting T24 cells with a cocktail of miR-31-5p, miR-29b-3p, and miR-126-3p inhibitors along with TSN siRNA or UPF1 siRNA restored invasion to ∼30%–45% of control knockdown cells (Fig. 5H,I; Supplemental Fig. S4C). Taken together, our data demonstrate that TSN and UPF1 promote T24 bladder cancer cell invasion at least in part by degrading anti-invasive miRNAs.
Discussion
The results shown here extend UPF1 helicase function beyond mRNA decay to the decay of mature miRNAs in a process that we named TumiD, adding a new layer to the roles of helicases in RNAi pathways (Figs. 1–5). Our results also extend the role of TumiD from promoting the G1-to-S-phase transition (Elbarbary et al. 2017) to promoting bladder cancer cell invasion (Fig. 5).
Precedents exist in which reorganization of RNA−protein complexes by the UPF1 helicase is essential for RNA decay, as exemplified by NMD and Regnase-1-mediated mRNA decay (Franks et al. 2010; Gregersen et al. 2014; Mino et al. 2015). In fact, UPF1 might function in TumiD to remodel RISC-bound miRNAs not only in complex with (Fig. 3D,E) but also free of target mRNAs (Fig. 6). The timing of UPF1 recruitment to miRNAs relative to UPF1 activation to dissociate miRNAs from their target mRNAs during TumiD remains to be determined. From our data, UPF1 and TSN, which at least in vitro interact directly (Fig. 1E), may be core components of RISC or recruited to RISC to initiate miRNA degradation (Fig. 6). While cellular UPF1 and TSN coimmunoprecipitate without cross-linking prior to cell lysis, neither UPF1 nor TSN coimmunoprecipitates with core RISC component AGO2 or GW182 in the absence of cross-linking (data not shown), which might suggest, among other scenarios, a transient interaction. In the case where UPF1 and TSN are recruited to RISC, as one of many possibilities, methylation of one or more RISC components might promote TSN recruitment—and therefore TumiD—given that the Tudor domain of TSN is known to interact with methylated proteins (Chen et al. 2011). In either case, TSN and UPF1 may gain access to target miRNAs upon regulated RISC reorganization. Other helicases in addition to UPF1 are likely to be involved in recycling RISC, since UPF1 knockdown does not inhibit miRNA-mediated target silencing (Fig. 5E–G).
Figure 6.
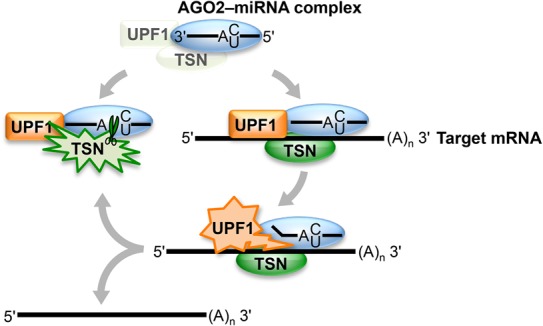
Model for TumiD, in which the complex of TSN–UPF1 might be a core component of RISC or recruited to RISC. Once TSN–UPF1 gains access to a miRNA, which may involve regulated RISC reorganization, UPF1 helicase activity frees the miRNA from its target mRNA, rendering the miRNA susceptible to TSN-mediated endonucleolytic cleavage at accessible CA and UA dinucleotides. Green starbursts denote TSN cleavage activity, orange starbursts denote UPF1 helicase activity, and blue ovals denote RISC (i.e., the AGO2–miRNA complex).
Our miR-seq data for T24 bladder cancer cells indicate that UPF1 and TSN mediate the decay of a vast repertoire of mature miRNAs (Fig. 4; Supplemental Table S3). Notably, the current number of TumiD targets is an underestimate due to our inability to down-regulate TSN or UPF1 for extended periods, which would inhibit the cell cycle and cell proliferation (Azzalin and Lingner 2006; Cappellari et al. 2014; Lou et al. 2014; Su et al. 2015; Elbarbary et al. 2017).
Decreased expression of miR-31-5p is associated with aggressive tumor progression and poor prognosis for different cancer types that include breast and bladder cancers (Augoff et al. 2011; Wang et al. 2013). While the expression of miR-31-5p is epigenetically silenced in some invasive breast cancer cell lines, it is not in others, indicating that there can be regulation of miR-31-5p expression via nonepigenetic mechanisms (Valastyan and Weinberg 2010). In the present study, we present TumiD as a novel means to regulate the level of mature miR-31-5p in cancer cells.
TumiD is undoubtedly important to many other biological processes. For example, it will be interesting to test whether ADAR2-mediated miRNA editing at TSN targeted CA and/or UA dinucleotide cleavage sites inhibits TumiD. Underscoring this prospect, 70% of the ADAR2-mediated A-to-I-editing events occurring within mature miRNAs of mouse brains, which manifests a miRNA-editing level of >5%, modify A residues within CA and UA dinucleotides; of these miRNAs, 14% are down-regulated, possibly by TumiD, upon ADAR2 knockout (Vesely et al. 2014).
Materials and methods
Raw data were deposited in the Gene Expression Omnibus (accession no. GSE100068).
Plasmid constructions
pCMV-MYC-UPF1R(1–1118), pCMV-MYC-UPF1R (R843C), pCMV-MYC, pCMV-3xFlag-TSNR, pET28a-EGFP, pET28a-TSN(1–885), and pLHCX-3xFlag-AGO2R (used to establish HEK293T cells that stably express 3xFlag-AGO2R) were described previously (Kurosaki et al. 2014; Elbarbary et al. 2017). The remaining TSN- and UPF1-expressing plasmids were siRNA-resistant (R) but are referred to here without the superscript designation unless it was relevant to the experiment.
To construct the retroviral expression vector pLHCX-MYC-UPF1R(1–1118), the entire MYC-UPF1R(1–1118)-coding region was excised from pCMV-MYC-UPF1R(1–1118) using SacII. The SacII-generated ends were made blunt using Quick Blunting kit (BioLabs), and the resulting blunt-ended fragment was inserted into HpaI-digested pLHCX (Clontech).
To construct the retroviral expression vector pLHCX-MYC, the synthetic DNA duplex of 5′-ATGGCATCAATGCAGAAGCTGATCTCAGAGGAGGACCTGTAA-3′ and 5′-TTACAGGTCCTCCTCTGAGATCAGCTTCTGCATTGATGCCAT-3′ was inserted into HpaI-digested pLHCX (Clontech).
To construct the lentiviral expression vector pLVX-Puro-3xFlag-TSNR, the entire 3xFlag-TSNR sequence was PCR-amplified using pCMV-3xFlag-TSNR and the primer pair 5′-AGCGTTAACATGGACTACAAAGACCATGACGG-3′ (sense, where underlined nucleotides specify a HpaI site) and 5′-AGCGTTAACT TAGCGGCTGTAGCCAAATTCG-3′ (antisense, where underlined nucleotides specify a HpaI site). The resulting PCR product was digested using HpaI and ligated to SmaI-digested pLVX-Puro (Clontech).
To construct the retroviral expression vector pLVX-Puro-3xFlag, the synthetic DNA duplex of 5′-ATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACTAA-3′ and 5′-TTAGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCCAT-3′ was inserted into SmaI-digested pLVX-Puro (Clontech).
Cell culture and transfections
Human HEK293T and Dicer knockdown 2b2 (Schmitter et al. 2006) cells were propagated in Dulbecco's modified Eagle's medium (Gibco), and T24 cells were propagated in RPMI-1640 (Gibco). Both media were supplemented with 10% fetal bovine serum. Where specified, cells were transfected with 50 nM siRNA (GE Healthcare Dharmacon), 10 nM miRNA mimic (Qiagen), or 10 nM miRNA inhibitor (Qiagen) using RNAiMAX (Invitrogen).
HEK293T cells stably expressing Flag-TSN + MYC-UPF1 or, as controls, Flag or MYC were generated using pLHCX-MYC, pLHCX-MYC-UPF1R(1–1118), pLVX-Puro-3xFlag, or pLVX-Puro-3xFlag-TSNR as detailed (Popp and Maquat 2015). HEK293T cells stably expressing 3xFlag-AGO2R have been described (Elbarbary et al. 2017). To induce Dicer shRNA expression, Dicer knockdown 2b2 cells were cultured in 1 µg/mL doxycycline hydrochloride (Sigma). Notably, superscript “R,” indicating siRNA resistance, is omitted from figures and the text when not relevant to experiments.
Immunoprecipitations
Cells were cross-linked using 0.2% formaldehyde (Sigma) prior to lysis, and lysate proteins and/or RNAs were analyzed before and after immunoprecipitation as reported (Gong et al. 2009) with slight modifications: Cells were lysed, and immunoprecipitations were performed in lysis buffer (20 mM HEPES at pH 7.4, 300 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 1 mM dithiothreitol [DTT], 0.5% NP-40, Complete mini, EDTA-free protease inhibitor cocktail tablets [Roche]), and, when indicated, 1 U/µL RNase I (Promega) was used for RNA digestion as described (Singh et al. 2012). Immunoprecipitations of MYC, TSN, and UPF1 were performed using Dynabeads protein G/A (Life Technologies) conjugated with, respectively, monoclonal anti-c-MYC (Calbiochem), anti-TSN (Abcam), and anti-UPF1 (Serin et al. 2001), while immunoprecipitations of Flag were performed using anti-Flag M2 agarose affinity gel (Sigma).
Mass spectrometry
3xFlag-TSN or MYC-UPF1 was immunoprecipitated from HEK293T cells stably expressing 3xFlag and MYC or 3xFlag-TSN and MYC-UPF1 using anti-Flag (Sigma) or anti-c-MYC (Calbiochem) in the presence of RNase I. For mass spectrometric analysis, immunoprecipitates were resolved in SDS–polyacrylamide using 4%–15% Mini-Protean TGX gels (Bio-Rad), and bands detected using SilverQuest (Invitrogen) were excised and subjected to in-gel trypsin digestion (Kinter and Sherman 2000). Chromatographed peptides were identified by Eric Spooner at the Whitehead Institute Mass Spectrometry Facility (http://jura.wi.mit.edu/massspec/home.html) using an Orbitrap Elite Hybrid Ion Trap-Orbitap mass spectrometer with Dionex Ultimate 3000 Rapid Separation LC systems (Thermo Fisher Scientific) and Scaffold (version Scaffold_4.2.1, Proteome Software, Inc.).
Western blotting
Cells were lysed in RIPA buffer (Sefton 2001), and total cell proteins were separated in a 6%–15% denaturing polyacrylamide gel, transferred to nitrocellulose (Amersham Biosciences), and probed using antibodies recognizing TSN (Abcam), UPF1 (Serin et al. 2001), Flag (Sigma), MYC (Calbiochem), AGO2 (Abcam), GW182 (Bethyl Laboratories), PABPC1 (Santa Cruz Biotechnology), calnexin (Enzo Life Sciences), HIS (Qiagen), GAPDH (Santa Cruz Biotechnolgy), ITGA5 (Abcam), ITGA2 (Abcam), ANGPTL4 (Thermo Fisher Scientific), ADAM9 (Abcam), or Dicer (Abcam). Blots were quantified using Image Quant (Molecular Dynamics).
Purification of recombinant proteins
HIS-tagged TSN variants and HIS-tagged EGFP were expressed in BL21-CodonPlus(DE3)-RIPL cells (Agilent Technologies), purified using a HisTrap HP column (GE Healthcare), and further purified on a HiTrap Q HP column (GE Healthcare) as described (Elbarbary et al. 2017). Flag-UPF1 was expressed in Spodoptera frugiperda Sf9 cells, immunoprecipitated and eluted as described (Bhattacharya et al. 2000; Hosoda et al. 2005), purified using a HiTrap Q HP column, and dialyzed in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM DTT. UPF1(115–915) wild type and UPF1(115–915)R843C were expressed and purified as described (Kurosaki et al. 2014). Concentrations of recombinant proteins were estimated after electrophoresis in SDS–polyacrylamide using Coomassie blue (Sigma), where serial dilutions of bovine serum albumin (Rockland) provided concentration standards.
In vitro pull-down assays
Ten picomoles of E. coli-produced HIS-EGFP or HIS-TSN(1–885) was incubated with baculovirus-produced Flag-UPF1 in HIS pull-down buffer (30 mM Tris-HCl at pH 8.0, 100 mM NaCl, 10% [v/v] glycerol, 0.1% Triton X-100, 1 mM DTT, Complete mini, EDTA-free protease inhibitor cocktail tablets [Roche]) for 16 h at 4°C. The mixture then was incubated for 2 h at 4°C with Flag-peptide-saturated HisPur Ni-NTA resin. Resin was generated by incubating 50 µL of HisPur Ni-NTA resin slurry (Pierce) with 10 µg of Flag-peptide (Sigma) in HIS pull-down buffer for 2 h at 4°C and was subsequently washed five times using the same buffer. Mixture-incubated resin was washed 10 times in HIS pull-down buffer. Bound proteins were eluted by boiling in 2× SDS sample buffer (125 mM Tris-HCl at pH 6.8, 4% [w/v] SDS, 10% [w/v] 2-mercaptoethanol, 0.01% [w/v] Bromophenol blue), electrophoresed in SDS–polyacrylamide, and subjected to Western blotting using anti-Flag (Sigma) or anti-HIS (Qiagen).
siRNAs
Control siRNA, TSN siRNA (Elbarbary et al. 2017), and UPF1 siRNA (Kim et al. 2005) have been described.
RNA purification and RT-qPCR
RNA purification and RT-qPCR were performed as described previously (Elbarbary et al. 2017).
Tandem immunoprecipitations
HEK293T cells stably expressing Flag-TSN and MYC-UPF1 were formaldehyde cross-linked, lysed, and immunoprecipitated using the anti-Flag affinity gel as described above. The beads were washed five times with ice-cold lysis buffer and then twice with wash buffer (20 mM HEPES at pH 7.4, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 1 mM DTT, Complete mini, EDTA-free protease inhibitor cocktail [Roche]) followed by incubation with RNase I as described above. After an additional five washes with lysis buffer, Flag-TSN-containing complexes were affinity-eluted from the beads with elution buffer (200 µg/mL Flag peptide [Sigma], 5% glycerol in wash buffer) with rocking overnight at 4°C. NaCl and NP-40 were adjusted to 300 mM and 0.5%, respectively, and the second immunoprecipitation was performed for 2 h at 4°C using anti-MYC that had been coupled to Dynabeads protein G (Life Technologies) according to the manufacturer's instructions. Beads were washed seven times using lysis buffer, and complexes were eluted using 2× SDS sample buffer and decross-linked as reported (Gong et al. 2009).
In vitro nuclease and helicase assays
Synthetic single-stranded miRNAs (Integrated DNA Technology) were 5′ end-labeled using γ-[32P]ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs). For nuclease assays, 100–200 fmol of labeled miRNAs was incubated for 2 h at 37°C (unless otherwise indicated) with 1–2 pmol of E. coli-produced HIS-TSN(1–885) in 15 µL of nuclease reaction buffer (50 mM HEPES-KOH at pH 7.5, 5 mM CaCl2, 100 mM NaCl, 1 mM ATP). Reactions were terminated by adding 1 µL of proteinase K (Thermo Fisher Scientific) for 5 min at 37°C, and products were purified using RNeasy MinElute cleanup kits (Qiagen) or directly boiled in-gel loading buffer II (Thermo Fisher Scientific), electrophoresed in 6 M urea–15% polyacrylamide, and visualized using a Typhoon PhosphorImager (GE Healthcare). Helicase assays were performed by incubating duplexes for 2 h at 37°C with recombinant Flag-UPF1, UPF1(115–915) wild type, or UPF1(115–915)R843C in helicase buffer as described previously (Park et al. 2013).
Synthesizing the FZD3 mRNA 3′ UTR
The 80 nt of FZD3 mRNA 3′ UTR containing the miR-31-5p target site were synthesized in vitro using DNA generated by annealing sense (5′-TAATACGACTCACTATAGGGTCTGTATTCATTGTTGTATTTTTCTACAGTGAGATGTGATCTTGCCAAAGCCACCAGACCTTGGCTTCCAGGCCCTCCTG-3′) and antisense (5′-CAGGAGGGCCTGGAAGCCAAGGTCTGGTGGCTTTGGCAAGATCACATCTCACTGTAGAAAAATACAACAATGAATACAGACCCTATAGTGAGTCGTATTA-3′) strands and the T7 MEGAscript kit (Thermo Fisher Scientific). Product RNA was purified using RNeasy Minelute cleanup kits (Qiagen).
Assays of Flag-UPF1-mediated dissociation of target mRNA from AGO2-bound miRNA:target mRNA duplexes
Unlabeled miR-31-5p was loaded into AGO2 complexes in cell lysates as described previously (Elbarbary et al. 2017). The beads were further washed with loading buffer and subsequently incubated with internally [32P]-labeled FZD3 3′ UTR for 2 h at 37°C in loading buffer. Beads were then washed with lysis buffer until the final wash was free of radioactivity. Complexes were eluted from beads by incubation overnight at 4°C with helicase buffer that contained 150 µg/mL Flag peptide (Sigma). Eluted complexes were incubated for 2 h at 37°C with or without Flag-UPF1 as described previously (Park et al. 2013) and electrophoresed in a 1% native agarose gel.
Small RNA library preparation for Illumina sequencing
The TruSeq small RNA library preparation protocol (Illumina) was used to generate cDNA libraries using small RNA fractions (<200 nt) derived from T24 human bladder cancer cells. After addition of the TruSeq 3′ adapter (5′ adenylated) using truncated T4 RNA ligase II (New England Biolabs) and, subsequently, the 5′ adapter using T4 RNA ligase I (New England Biolabs), RNA was reverse-transcribed using a 3′ adapter-specific primer. The resulting cDNA was then amplified using 12 cycles of PCR, a universal forward primer, and a reverse primer harboring a bar code. cDNA libraries were purified using Pippin Prep (Sage Science) and quantitated first using an Agilent Bioanalyzer and subsequently using qPCR. Barcoded libraries were mixed at equimolar concentrations, and single-end 50-nt sequencing was undertaken using a HISeq2500v4 sequencer (Illumina). All steps were performed at the University of Rochester Genomics Research Center.
Computational analyses of miR-seq data
Raw reads generated from the Illumina HiSeq2500 sequencer were demultiplexed using configurebcl2fastq.pl version 1.8.4. miRge (Baras et al. 2015), in conjunction with cutAdapt 1.12 (Martin 2011) and Bowtie 1.2 (Langmead et al. 2009), was used to trim adapter sequences, align data to the miRge human miRNA annotation (built from mirBase version 21), and quantify miRNA reads for each sample with the following parameters: “--diff-isomirs --adapter illumina --species human.” Notably, the isoMir Bowtie alignment parameter setting was changed from “-n 2” to “-n 0” to more strictly define a read as an isoMir of a canonical miRNA sequence. Differential expression analyses were performed using the DESeq2 1.12.4 (Love et al. 2014) R/Bioconductor package (Huber et al. 2015) with an adjusted P-value (Benjamini-Hochberg) threshold of 0.05 within the R version 3.3.0 environment (https://www.R-project.org). Heat maps were produced using the pHeatmap version 1.0.8 (https://CRAN.R-project.org/package=pheatmap) package with row scaling and hierarchical clustering of the rLog transformed expression values. Correlation matrices showing Pearsons's correlation were created using the pair_corr() function from the pcaExplorer-2.0.0 (https://github.com/federicomarini/pcaExplorer) R/Bioconductor package. Significance values for the intersection of two miRNA lists were derived using the hypergeometric test (https://www.R-project.org).
T24 cell invasion assays
Three days after transfection with the specified siRNA or miRNA mimic, 4.0 × 104 T24 cells were seeded in BD BioCoat Matrigel invasion chambers with 8.0-mm pores (BD Biosciences) according to the manufacturer's instructions. After 22 h, invading cells were fixed using 100% methanol, stained using 1% (v/v) toluidine blue in 1% (w/v) borax, photographed using an Axiovert 40 CFL microscope (Zeiss), and quantitated using ImageJ (http://www.nih.gov).
Proliferation assays
Assays were performed using the CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer's instructions.
Supplementary Material
Acknowledgments
We thank R. Kallicharan-Smith, F. Lopez, and G. Pavlencheva for plasmid preparations; C. Beckham for T24 cells; and M. Popp for comments on the manuscript. This work was supported by National Institutes of Health (NIH) R37 GM74593 to L.E.M., NIH Pilot Award P30 AR061307 to R.A.E., and the Uehara Memorial Foundation to K.M.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.303537.117.
References
- Ambrus AM, Frolov MV. 2009. The diverse roles of RNA helicases in RNAi. Cell Cycle 8: 3500–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. 2011. miR-31 is a broad regulator of β1-integrin expression and function in cancer cells. Mol Cancer Res 9: 1500–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J. 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16: 433–439. [DOI] [PubMed] [Google Scholar]
- Backes C, Khaleeq QT, Meese E, Keller A. 2016. miEAA: microRNA enrichment analysis and annotation. Nucleic Acids Res 44: W110–W116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baras AS, Mitchell CJ, Myers JR, Gupta S, Weng LC, Ashton JM, Cornish TC, Pandey A, Halushka MK. 2015. miRge—A multiplexed method of processing small RNA-Seq data to determine microRNA entropy. PLoS One 10: e0143066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CF, Mortreux F, Auboeuf D. 2016. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Biol 17: 426–438. [DOI] [PubMed] [Google Scholar]
- Cappellari M, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O, Sette C. 2014. The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene 33: 3794–3802. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414. [DOI] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, Pawson T. 2011. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol 12: 629–642. [DOI] [PubMed] [Google Scholar]
- Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. 2013. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol 15: 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Franks TM, Lykke-Andersen J. 2016. Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat Commun 7: 12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Miyoshi K, Meyers JR, Du P, Ashton JM, Tian B, Maquat LE. 2017. Tudor-SN-mediated endonucleolytic decay of human-cell microRNAs promotes G1/S phase transition. Science 356: 859–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini F, Bagchi D, Le Hir H, Croquette V. 2015. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat Commun 6: 7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143: 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. 2009. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 23: 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. 2014. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54: 573–585. [DOI] [PubMed] [Google Scholar]
- Gurtan AM, Sharp PA. 2013. The role of miRNAs in regulating gene expression networks. J Mol Biol 425: 3582–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Hammond SM. 2005. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett 579: 5822–5829. [DOI] [PubMed] [Google Scholar]
- Hosoda N, Kim YK, Lejeune F, Maquat LE. 2005. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 12: 893–901. [DOI] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. 2015. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 25: 651–665. [DOI] [PubMed] [Google Scholar]
- Jariwala N, Rajasekaran D, Srivastava J, Gredler R, Akiel MA, Robertson CL, Emdad L, Fisher PB, Sarkar D. 2015. Role of the staphylococcal nuclease and tudor domain containing 1 in oncogenesis. Int J Oncol 46: 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia AY, Castillo-Martin M, Bonal DM, Sánchez-Carbayo M, Silva JM, Cordon-Cardo C. 2014. MicroRNA-126 inhibits invasion in bladder cancer via regulation of ADAM9. Br J Cancer 110: 2945–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Suh MR, Han J, Yeom KH, Lee Y, Heo I, Ha M, Hyun S, Kim VN. 2009. Human UPF1 participates in small RNA-induced mRNA downregulation. Mol Cell Biol 29: 5789–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karousis ED, Nasif S, Mühlemann O. 2016. Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA 7: 661–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208. [DOI] [PubMed] [Google Scholar]
- Kinter M, Sherman NE. 2000. Protein sequencing and identification using tandem mass spectrometry. John Wiley Sons, Inc, New York. [Google Scholar]
- Kurosaki T, Maquat LE. 2016. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129: 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, Tian B, Maquat LE. 2014. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev 28: 1900–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Pratt GA, Martinez FJ, Yeo GW, Lykke-Andersen J. 2015. Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol Cell 59: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RG, Supek F, Lehner B. 2016. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet 48: 1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. 2014. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep 6: 748–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal 17: 10–12. [Google Scholar]
- Michlewski G, Cáceres JF. 2010. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol 17: 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels HH, Ori D, Uehata T, Tartey S, Akira S, Suzuki Y, et al. 2015. Regnase-1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161: 1058–1073. [DOI] [PubMed] [Google Scholar]
- Ottens F, Gehring NH. 2016. Physiological and pathophysiological role of nonsense-mediated mRNA decay. Pflugers Arch 468: 1013–1028. [DOI] [PubMed] [Google Scholar]
- Park E, Maquat LE. 2013. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA 4: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Gleghorn ML, Maquat LE. 2013. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci 110: 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. 2015. Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat Commun 6: 6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. 2016. Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell 165: 1319–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD. 2005. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol 12: 489–496. [DOI] [PubMed] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. 2006. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res 34: 4801–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefton BM. 2001. Labeling cultured cells with 32Pi and preparing cell lysates for immunoprecipitation. Curr Protoc Cell Biol 3: 14.4.1–14.4.7. [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Zhang C, Tecle A, Fu X, He J, Song J, Zhang W, Sun X, Ren Y, Silvennoinen O, et al. 2015. Tudor staphylococcal nuclease (Tudor-SN), a novel regulator facilitating G1/S phase transition, acting as a co-activator of E2F-1 in cell cycle regulation. J Biol Chem 290: 7208–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. 2010. miR-31: a crucial overseer of tumor metastasis and other emerging roles. Cell Cycle 9: 2124–2129. [DOI] [PubMed] [Google Scholar]
- Vesely C, Tauber S, Sedlazeck FJ, Tajaddod M, von Haeseler A, Jantsch MF. 2014. ADAR2 induces reproducible changes in sequence and abundance of mature microRNAs in the mouse brain. Nucleic Acids Res 42: 12155–12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD, Prionidis K, Dalamagas T, Hatzigeorgiou AG. 2012. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res 40: W498–W504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li Q, Wang K, Dai Y, Yang J, Xue S, Han F, Zhang Q, Liu J, Wu W. 2013. Decreased expression of microRNA-31 associates with aggressive tumor progression and poor prognosis in patients with bladder cancer. Clin Transl Oncol 15: 849–854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.