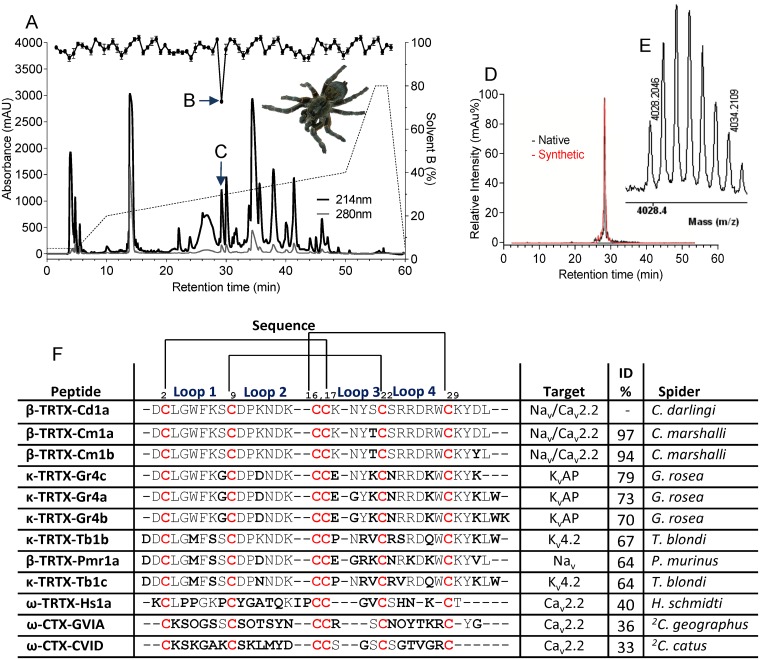
Fig 1. Assay-guided isolation of Cd1a.
Assay-guided fractionation of Cd1a from C. darlingi venom. (A) Crude venom (0.5 mg/100 μL) was injected onto a RP-HPLC column (Vydac C18) and fractionated using a linear gradient of solvent B: 5% for 5 min, 5–20% for another 5 min and 20–40% over 40 min reaching 80% at 50–60 min. Fraction corresponding to Cd1a eluted with a retention time of 28 min corresponding to ~30% solvent B (indicated by vertical arrow). This fraction fully inhibited KCl/CaCl2-evoked Cav2.2 responses (indicated by black circles over the chromatogram and a horizontal arrow) in SH-SY5Y cells, as measured by fluorescence-based Ca2+ imaging assays. (D) Analytical HPLC traces for native (black) and synthetic (red) Cd1a show identical retention time and peak width. MALDI-TOF analyses. The observed mass of native Cd1a (M+H: 4028.2 Da) was consistent with that predicted from the amidated Edman-derived amino acid sequence. (F) Sequence alignment of Cd1a with similar spider peptides and peptidic Cav2.2 inhibitors (ω-toxins) from spider and cone snail venoms. Sequences were manually aligned. Non-conserved residues (compared to Cd1a) are highlighted in bold while the conserved cysteine framework is shown in red. The location of the four intercystine loops and the disulfide connectivity pattern are shown above the alignment. Abbreviations: Spider/cone snail genus: C. = Ceratogyrus, G. = Grammostola, T. = Theraphosa, P. = Pterinochilus, H. = Haplopelma, 2C. = Conus.