Abstract
Objectives
Pregnancy characteristics may influence the infant fecal microbiota during early life. We aimed to examine associations of maternal gestational weight gain with infant fecal microbiota composition, bacterial community richness, and Shannon diversity index.
Methods
We analyzed data from a prospective cohort study of healthy infants. We collected prenatal data, including report of mother’s gestational weight gain, and infant fecal samples from 84 infant-mother dyads. By applying 16S rRNA gene sequencing and an unbiased clustering by partitioning around medoids using Bray-Curtis distances, we identified four fecal microbiota profiles, and examined the associations of maternal gestational weight gain with the four fecal microbiota profiles, bacterial community richness, and Shannon diversity index.
Results
Overall, the median age of infants was 4.0 months and 43% were female. The mothers of the 84 infants gained a mean of 14.2 kg (SD, 5.4 kg) during pregnancy. We identified four distinct microbiota profiles: Bifidobacterium-dominant (42%), Enterobacter/Veillonella-dominant (23%), Bacteroides-dominant (19%), and Escherichia-dominant (17%). Infants whose mothers had higher gestational weight gain were less likely to have a Bacteroides-dominant profile, corresponding to a relative risk ratio of 0.83 (95%CI, 0.71–0.96; P=0.01) per 1 kilogram increase in weight. Additionally, higher gestational weight gain was also associated with lower bacterial community richness and Shannon diversity index (P<0.05).
Conclusion
In this prospective cohort study of healthy infants, maternal gestational weight gain was associated with the infant fecal microbiota profiles, bacterial community richness, and Shannon diversity index.
Introduction
The human intestine is colonized by trillions of commensal bacteria that serve varied physiologic functions, ranging from digestion of nutrients1 to immune development and modulation2,3. Recent evidence suggests that certain fecal microbiota in childhood can contribute to the pathogenesis of various disease states including obesity4,5, diabetes5, and asthma6. Over the first three years of life, the structure and composition of the fecal microbiota evolves to a more stable form7.
Emerging evidence indicates that the fecal microbiota during infancy is shaped by several prenatal and postpartum environmental factors8. Previous studies have found differences, at least in the short-term, in the infant fecal microbiota based on mode of delivery, initiation and duration of breastfeeding, and infant exposure to antibiotics8,9. Another factor that may affect the infant microbiota is maternal weight gain during pregnancy. Gestational weight gain (GWG) appears to be associated with several child outcomes including the development of asthma10 as well as obesity and adiposity11. However, few studies have directly examined the relationship between GWG and composition of the infant microbiome12.
To address this knowledge gap, by examining the fecal microbiota in a prospective cohort of healthy infants, we aimed to determine associations of maternal GWG with infant fecal microbiota profiles, bacterial community richness, and Shannon diversity index.
Methods
Study Design and Participants
We analyzed data from a prospective cohort study of healthy infants. The study design, setting, participants, and methods of data collection have been reported previously13,14. Briefly, as part of the 43rd Multicenter Airway Research Collaboration (MARC-43)15, a multicenter prospective cohort study, we enrolled 120 healthy infants at Massachusetts General Hospital (Boston, MA) from November 2013 through April 2014. Inclusion criteria were age <1 year and gestational age ≥34 weeks at birth. Infants were excluded if they had comorbidities (heart-lung disease, immunodeficiency, immunosuppression, or chronic gastrointestinal disorder), had fever, respiratory illness, or gastrointestinal illness, or had been treated with antibiotics in the 7 days prior to enrollment. This study was approved by the Partners institutional review board. Written informed consent was obtained from the parent or legal guardian.
Data and Fecal Sample Collection
Site investigators conducted structured interviews and medical record reviews to collect information on maternal and child demographic characteristics (e.g., age, sex, race/ethnicity), family history, prenatal and past medical history (e.g., mode of birth, gestational age, systemic antibiotic use), and nutritional/environmental characteristics (mode of feeding, daycare attendance) at enrollment and 2 years of age.
Infants’ fecal samples were collected by parents at home using a standardized protocol13,14. First, parents were instructed to collect diapers from their infant, which contained feces, and immediately refrigerate them. Within 24 hours, stool was returned to the study staff and immediately placed in sterile Sarstedt feces collection containers (Sarstedt; Nümbrecht, Germany) and stored at −80°C. Frozen samples were shipped on dry ice to the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine, where samples were processed and the microbiomes were characterized using 16S rRNA gene sequencing.
Primary Exposure
The primary exposure was maternal GWG. To assess GWG, parents were asked: how much weight did the [child]’s biological mother gain when she was pregnant? Based on previous literature11,16 and the distribution of our analytic sample (n=84), we divided GWG into three groups: ≤11.9 kg, 12.0–14.9 kg, and ≥15.0 kg.
Outcome Measures
The primary outcome in this study was infant fecal microbiota profile. Secondary outcomes were fecal bacterial community richness (the number of operational taxonomic units [OTUs]) and the Shannon diversity index score. Shannon index is a measure of within-sample (alpha) diversity based on the sum of the proportion of each species relative to the total number of species in the community and therefore accounts for both abundance and evenness.17 To examine the composition of the fecal microbiota, we applied a 16S rRNA gene sequencing approach. Detailed protocol for 16S rRNA gene sequencing has been described previously18 and was based on methods developed for the National Institutes of Health Human Microbiome Project19. In brief, bacterial genomic DNA was extracted using MO BIO PowerMag DNA Isolation Kit (Mo Bio Lab; Carlsbad, CA). The 16S rDNA V4 region was amplified by PCR and sequenced in the MiSeq platform (Illumina, SanDiego, CA) using 2×250 bp paired-end protocol. Sequencing read pairs were demultiplexed based on the unique molecular barcodes, and reads were merged using USEARCH v7.0.1090.20 16S rRNA gene sequences were clustered into operational taxonomic units (OTUs) at a similarity cutoff value of 97% using the UPARSE algorithm;21 OTUs were mapped to the SILVA database to determine taxonomies.22 Abundances were recovered by mapping the demultiplexed reads to the UPARSE OTUs.
Quality Control
The microbial DNA extraction, 16S rRNA gene amplification, and amplicon sequencing used a set of controls to evaluate the potential introduction of contamination. Non-template controls (extraction chemistries) were included in the microbial DNA extraction process and the resulting material was subsequently used for polymerase chain reaction (PCR) amplification. Additionally, at this step, to evaluate the potential introduction of contamination, another set of non-template controls (PCR-mix) was included. Similarly, to evaluate the efficiency of the amplification process, a positive control comprised of known and previously characterized microbial DNA was included. Before samples (unknowns) are pooled together, sequencing controls were evaluated, and the rejection criteria were the presence of amplicons in any of the non-template controls or the absence of amplicons in the positive control. As another level of quality control, both non-template and positive controls were included in the sequencing pool. The resulting sequence reads associated with the controls were examined. The acceptance criteria were an alignment of >99% of alignment of the sequence reads of the positive control to the expected bacterial genome, and <500 raw sequence reads in the non-template controls. In the current study, no amplicons were observed in the non-template controls and a negligible amount of reads were recovered after sequencing.
Statistical Analysis
We conducted microbiota analyses at the genus-level because the bacterial DNA sequences in our sample were dominated by one OTU per genus18. To characterize the fecal microbiota profiles, we performed unbiased clustering by partitioning around medoids (PAM)23 using Bray-Curtis distances. The number of clusters chosen for the data was determined using the gap statistic.24
Next, we examined the associations of maternal GWG with infant microbiota profiles by constructing unadjusted and adjusted multinomial regression models with fecal microbiota profiles as the independent variable. We constructed adjusted models controlling for different sets of potential confounders: infant age (at stool collection), household income, delivery mode, lifetime history of systemic antibiotic use, and mode of feeding (breastfed vs. formula-fed). These variables were chosen based on a priori knowledge of differences in microbiota composition.8,25 Given the relatively small number of infants, we included ≤2 covariates per model. We then determined if the relationship between GWG and fecal microbiota profiles was modified by infant feeding status; that is, did infant microbiota profiles vary by GWG within the group of breast-fed infants (n=66) and within the group of formula-fed infants (n=18). In addition, to examine the association of maternal GWG with fecal bacterial community richness (number of genera) and Shannon diversity index, we used Spearman’s correlation and Pearson’s correlation, respectively. We conducted statistical analyses using R version 3.3. A two-tailed P<0.05 was considered statistically significant.
Results
Study Population
Of 115 enrolled infants with fecal samples, we excluded 31 infants without maternal GWG data, leaving 84 (73%) eligible for the present analysis. The analytic and nonanalytic cohorts were similar in patient age, sex, race/ethnicity, prenatal and past medical history, and environmental characteristics (all P>0.05). In contrast, the analytic cohort significantly differed from the non-analytic cohort with regard to annual household income, infant systemic antibiotic use, mode of feeding, and alpha diversity index of fecal microbiota (all P<0.05) (Supplemental Digital Content, Table 1: Patient and Fecal Microbiota Characteristics in the Analytic and non-Analytic Cohort).
Overall, the median age of infants was 4.0 months (IQR 2.0–4.7), 43% were female, and 52% were non-Hispanic white. The mothers of the 84 infants in the analytic cohort gained a mean of 14.2 kg (SD, 5.4 kg) during pregnancy. Table 1 presents the infant and microbiota characteristics by GWG group. Most subject characteristics did not differ significantly across the GWG groups (all P>0.05), although maternal GWG was related to primarily breastfeeding during the first 3 months postpartum (P=0.04).
Table 1.
Subject and Fecal Microbiota Characteristics in Infants According to Gestational Weight Gain Category (n=84)
| Characteristics | Maternal gestational weight gain | P-value |
||
|---|---|---|---|---|
| ≤11.9 kg n=26 |
12.0–14.9
kg n=24 |
≥15.0 kg n=34 |
||
| Subject characteristics | ||||
| Demographics | ||||
| Age (mo), median (IQR) | 4.1 (2.2–4.6) | 3.7 (2.1–4.8) | 2.1 (1.8–4.9) | 0.36 |
| Female sex | 9 (35) | 13 (54) | 14 (41) | 0.37 |
| Race/ethnicity | 0.20 | |||
| Non-Hispanic white | 15 (58) | 15 (62) | 14 (41) | |
| Non-Hispanic black | 4 (15) | 1 (4) | 3 (9) | |
| Hispanic | 5 (19) | 3 (12) | 5 (15) | |
| Other | 2 (8) | 5 (21) | 12 (35) | |
| Annual household income | 0.06 | |||
| <$40,000 | 4 (16) | 1 (4) | 2 (6) | |
| $40,000 to $99,999 | 7 (28) | 13 (53) | 9 (27) | |
| $100,000 or more | 15 (58) | 10 (42) | 23 (68) | |
| Prenatal history | ||||
| Maternal age (year), median (IQR) | 34 (31–37) | 32 (29–37) | 31 (28–36) | 0.23 |
| Maternal smoking during pregnancy | 1 (4) | 0 (0) | 0 (0) | 0.32 |
| Maternal antibiotic use during pregnancy | 2 (8) | 3 (12) | 4 (12) | 0.83 |
| Maternal antibiotic use during labor | 11 (42) | 10 (42) | 6 (18) | 0.06 |
| Past medical history | ||||
| Mode of birth | 0.56 | |||
| Vaginal birth | 15 (58) | 17 (71) | 20 (59) | |
| Cesarean section | 11 (42) | 7 (29) | 14 (41) | |
| Prematurity (32–37 weeks) | 2 (8) | 2 (8) | 6 (18) | 0.41 |
| History of systemic antibiotic use before enrollment | 4 (15) | 3 (12) | 4 (12) | 0.91 |
| Environmental characteristics | ||||
| Mostly breastfed for the first 3 months of age | 16 (62) | 21 (88) | 29 (85) | 0.04 |
| Ever attended daycare | 3 (12) | 3 (12) | 5 (15) | 0.93 |
| Fecal microbiota characteristics | ||||
| Microbiota profiles | 0.03 | |||
| Enterobacter-dominant profile | 3 (12) | 8 (33) | 8 (24) | |
| Escherichia-dominant profile | 1 (4) | 6 (25) | 7 (21) | |
| Bifidobacterium-dominant profile | 13 (50) | 6 (25) | 16 (47) | |
| Bacteroides-dominant profile | 9 (35) | 4 (17) | 3 (9) | |
| Number of genera, median (IQR) | 19 (13–23) | 13 (10–17) | 14 (10–18) | 0.01 |
| Shannon diversity index, mean (SD) | 1.64 (0.51) | 1.32 (0.44) | 1.30 (0.44) | 0.01 |
Data are no. (%) of infants unless otherwise indicated. Percentages may not equal 100 because of missingness or rounding
Abbreviations: mo, months; IQR, interquartile range; SD, standard deviation
Gestational Weight Gain and Infant Fecal microbiota
All 84 fecal samples had sufficient sequence depth to obtain high degree of sequence coverage (rarefaction cutoff, 1,470 reads per sample). The fecal microbiota was composed primarily of four genera – Escherichia (23%), Bifidobacterium (23%), Enterobacter (20%), and Bacteroides (12%). Unsupervised clustering of fecal microbiota identified 4 distinct microbiota profiles (Figure 1): 1) Bifidobacterium-dominant profile (42%), 2) Enterobacter/Veillonella-dominant profile (23%), 3) Bacteroides-dominant profile (19%), and 4) Escherichia-dominant profile (17%).
Figure 1. Principal coordinate analysis plot on infant fecal microbiota.
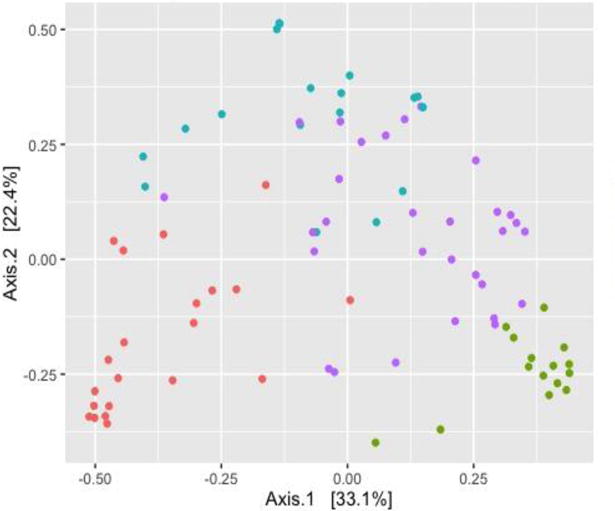
The Bray–Curtis distance between all subjects was calculated and used to generate principal coordinate analysis plot. Each dot in the figure represents the microbiota profile of a single subject in a two-dimensional space. Colored dots indicate 4 microbiota profiles: Escherichia-dominant profile (green), Bifidobacterium-dominant profile (blue), Enterobacter/Veillonella-dominant profile (red), and Bacteroides-dominant profile (purple). The subjects cluster together according to their microbiota profiles.
There was a negative linear relationship between the maternal GWG and likelihood of Bacteroides-dominant fecal microbiota profile in infants (P=0.01; Figure 2). In unadjusted multinomial models, higher GWG was associated with a decreased likelihood of a Bacteroides-dominant microbiota profile in infants (Table 2). Specifically, infants whose mothers had a higher GWG were less likely to have a Bacteroides-dominant profile than an Enterobacter-dominant profile, corresponding to an unadjusted relative risk ratio (RRR) of 0.83 (95%CI, 0.71–0.96; P=0.01) per 1 kg increase in weight. Although this relative risk was attenuated slightly, the associations remained significant in adjusted models (all P<0.05; Table 2).
Figure 2. Associations between maternal weight gain during pregnancy and proportion of Bacteroides-dominant fecal microbiota profile in infants.
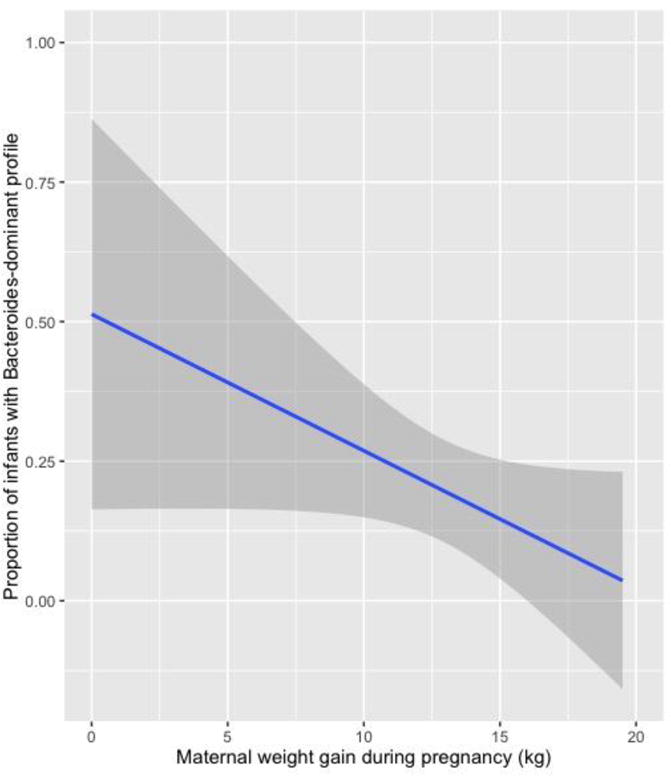
Association of maternal gestational weight gain and Bacteroides-dominant fecal microbiota profiles in infants. Infants whose mother had a greater gestational weight gain were less likely to have a Bacteroides-dominant profile, corresponding to an unadjusted relative risk ratio of 0.83 (95%CI, 0.71–0.96; P=0.01) per 1 kg increase. The grey shaded areas represent the 95% confidence interval.
Table 2.
Unadjusted and Adjusted Associations between Maternal Weight Gain During Pregnancy and Fecal Microbiota Profiles in Infants (n=84)
| Models and microbiota profiles | RRR* | 95% CI | P-value |
|---|---|---|---|
| Unadjusted model | |||
| Enterobacter-dominant profile | Reference | Reference | – |
| Escherichia-dominant profile | 0.97 | 0.84–1.11 | 0.61 |
| Bifidobacterium-dominant profile | 0.95 | 0.85–1.06 | 0.36 |
| Bacteroides-dominant profile | 0.83 | 0.71–0.96 | 0.01 |
| Adjusted model 1 (adjusted for age) | |||
| Enterobacter-dominant profile | Reference | Reference | – |
| Escherichia-dominant profile | 0.97 | 0.84–1.11 | 0.67 |
| Bifidobacterium-dominant profile | 0.96 | 0.85–1.08 | 0.47 |
| Bacteroides-dominant profile | 0.83 | 0.71–0.98 | 0.02 |
| Adjusted model 2 (adjusted for age and delivery mode) | |||
| Enterobacter-dominant profile | Reference | Reference | – |
| Escherichia-dominant profile | 0.97 | 0.85–1.12 | 0.72 |
| Bifidobacterium-dominant profile | 0.96 | 0.85–1.08 | 0.53 |
| Bacteroides-dominant profile | 0.83 | 0.70–0.98 | 0.03 |
| Adjusted model 3 (adjusted for age and history of systemic antibiotic use) | |||
| Enterobacter-dominant profile | Reference | Reference | – |
| Escherichia-dominant profile | 0.98 | 0.85–1.13 | 0.75 |
| Bifidobacterium-dominant profile | 0.96 | 0.85–1.09 | 0.54 |
| Bacteroides-dominant profile | 0.84 | 0.72–0.99 | 0.03 |
| Adjusted model 4 (adjusted for age and feeding status) | |||
| Enterobacter-dominant profile | Reference | Reference | – |
| Escherichia-dominant profile | 0.97 | 0.84–1.11 | 0.64 |
| Bifidobacterium-dominant profile | 0.97 | 0.86–1.09 | 0.61 |
| Bacteroides-dominant profile | 0.85 | 0.72–1.00 | 0.04 |
| Adjusted model 5 (adjusted for age and annual household income) | |||
| Enterobacter-dominant profile | Reference | Reference | – |
| Escherichia-dominant profile | 0.93 | 0.80–1.09 | 0.37 |
| Bifidobacterium-dominant profile | 0.92 | 0.81–1.06 | 0.25 |
| Bacteroides-dominant profile | 0.79 | 0.66–0.95 | 0.01 |
RRR, relative risk ratio; CI, confidence interval
RRR per 1 kg increase in maternal weight during pregnancy
We also examined if mode of feeding modified the association between GWG and microbiota profile. Although statistical power was limited, the stratified analysis suggested a non-significant association between GWG and likelihood of Bacteroides-dominant microbiota profile both in breast-fed (P=0.06) and formula-fed (P=0.21) infants (Supplemental Digital Content, Table 2: Unadjusted Associations between Gestational Weight Gain During Pregnancy and Fecal Microbiota Profiles in Infants, According to Feeding Status (n=84)).
Additionally, infant fecal bacterial community richness and Shannon diversity index score differed by GWG group (Table 1). Infant microbiota of mothers in the lowest GWG group had significantly greater bacterial community richness and greater Shannon diversity index compared to infant microbiota of mothers in the middle and highest GWG group (both P=0.01). Similarly, in the correlation analyses, there were negative correlations of GWG with bacterial community richness (Spearman’s rho=−0.25, P=0.02) and Shannon diversity index (Pearson’s rho=−0.25, P=0.02) (Supplemental Digital Content, Figure 1: Correlation of maternal weight gain during pregnancy with bacterial community richness and Shannon diversity index of infant fecal microbiota).
Discussion
The fecal microbiota of infants may be shaped by several factors during both the prenatal and postpartum periods8. In this analysis of a prospective cohort study of infants, we extend these prior findings by demonstrating that infants whose mothers had a higher GWG were less likely to have a Bacteroides-dominant microbiota profile. The association persisted after adjusting for different sets of potential confounders including child age at stool collection, delivery mode, exposure to antibiotics, and mode of feeding. In stratified analysis, mode of feeding did not significantly modify the association between GWG and microbiota profile. Additionally, we found that higher GWG was associated with lower bacterial community richness and a lower Shannon diversity index score of the infant fecal microbiota.
Prior studies have begun to explore the environmental factors during pregnancy and early infancy that may influence the child’s microbiome in early life (e.g., mode of delivery, exposure to antibiotics, and mode of infant feeding8,9). However, few have directly examined the relationship between maternal GWG and infant fecal microbiota12. In a smaller study with 42 Finnish mother-child pairs, Collado et al. used a quantitative PCR approach and found that the composition of infant fecal microbiota was related to both the mother’s pre-pregnancy weight and GWG. Specifically, higher GWG was associated with lower concentrations of Bacteroides and higher concentrations of Clostridium histolyticum in infants at one month of age. Similar findings, though statistically nonsignificant, were observed for both bacterial groups in infants at six months of age12. The current study, which relied on 16S rRNA gene sequencing, further supports the relationship between higher GWG and a lower likelihood of Bacteroides-dominant fecal microbiota in a larger prospective cohort of U.S. infants.
Prior studies have also observed associations between GWG and maternal microbiota, noting differences in several genera such as Clostridium, Escherichia, Bifidobacterium, and Bacteroides25,26. For example, Santacruz et al. found that normal weight gain during pregnancy is associated with increased abundances of Bacteroides, Bifidobacterium, and Akkermansia muciniphila in the maternal microbiota whereas excessive GWG is associated with increased abundances of Enterobacteriaceae and Escherichia coli26. Furthermore, studies examining the relationship between GWG and infant outcomes have found that higher GWG is an independent risk factor for the development of asthma10 and wheezing27, and increased BMI and adiposity11,28 in children. Taken together, these studies suggest that GWG may influence the fecal microbiota of both the mother and infant, as well as infant health outcomes.
The biological and metabolic significance of the intestinal microbiome during early childhood has been the subject of recent research. The literature suggest that Bacteroides serves vital functions including energy production1,29,30 and regulation of intestinal immunity2,3. In the human intestine, Bacteroides aid in the catabolism of complex polysaccharides which are ultimately broken down into short chain fatty acids which can be utilized as a significant source of energy by the host1,29,30. Additionally, Bacteroides species promote T cell and IL-10 production2,3. Depletion of Bacteroides in the intestine has been associated with development of asthma, allergies, and obesity31,32. Other studies, however, have found positive associations between abundance of Bacteroides and disease morbidity, including increased risk of obesity33 and bronchiolitis18. We also found that across GWG groups, Bifidobacterium-dominant profile was most common. Bifidobacterium have been previously shown to be the predominant microbiota in breast-fed infants34, consistent with the cohort of infants in the present study who were largely breast-fed. It is hypothesized that Bifidobacterium influence immune modulation as well as inflammatory status12,34.
Increased bacterial richness and diversity are also considered “protective”. For example, increased alpha diversity of infant fecal microbiota has been linked to decreased risk of type 1 diabetes35 and development of asthma6. Riva et al. found that the alpha diversity of fecal microbiota in children ages 6–16 years was negatively correlated with BMI z-score31. In our study, we found that higher GWG was associated with decreased bacterial community richness and decreased Shannon diversity index scores of infant fecal microbiota. By contrast, some studies have found greater fecal bacterial richness and diversity to be associated with increased odds of certain disease states (e.g., bronchiolitis, asthma)18,36. Inconsistencies among studies might be attributable to the differences in study design, population, methods for data collection and microbiome analysis, and outcome definitions – issues that are challenging to account for across studies29. Further research examining not only the microbiome structure, but also the metagenomics, metatranscriptomics, and metabolomics, is warranted to gain a more complete understanding of the functionalities of fecal microbiome and how they relate with long term health outcomes in children37.
The findings from the current study, along with previous studies, collectively suggest that maternal GWG may influence the composition and diversity of the infant fecal microbiota. However, the mechanism through which GWG affects the infant gut microbiome remains unclear. One possible explanation is that GWG may alter the microbiome of the mother which is ultimately transferred to the neonate. Indeed, emerging evidence has shown that weight gain during pregnancy is associated with differences in the composition of maternal fecal microbiota25,26 and that microbial transfer to offspring may begin in utero38.
Potential Limitations
There are several potential limitations of our study. First, maternal GWG was measured by self-report as opposed to independent observation, and thus there is a possibility of recall bias. Second, 27% of infants did not have maternal GWG data, and this absence is a potential source of bias. In particular, we found that despite offering mail-in survey collection and remuneration to families who completed the survey, the analytic and non-analytic cohorts differed by annual household income. We also did not have access to the mother’s pre-pregnancy BMI and were therefore unable to categorize GWG based on the Institute of Medicine’s BMI-specific recommendations for GWG39. However, even if we had this information we would have likely not had a large enough sample to permit this multi-level stratification. Third, as with any observational study, the observed associations do not prove causality. The relatively small sample size also precluded us from adjusting for all sets of potential confounders; however, the significant association persisted even after we controlled for clinically important covariates. Fourth, participants in this analysis were recruited from a single primary care clinic in an urban area; therefore, these results may not be generalizable to other populations. Finally, this study included infant microbiota data from the first year of life, so it is not known whether these differences in microbiota will persist in later childhood. To address this important question, the study cohort is being followed longitudinally to age 6 years with fecal specimen sampling at multiple time points.
It is important to note that although this study found that higher GWG was associated with a lower likelihood of Bacteroides-dominant microbiota profile, as well as lower bacterial community richness and Shannon diversity index, these findings are not meant to suggest that mothers should aim to dramatically limit or avoid weight gain during pregnancy. There is currently no known optimal weight gain during pregnancy, although recommendations range from approximately 10–20kg16. Some weight gain during pregnancy is important for the health of the developing fetus and should be encouraged.
In summary, based on the data from a prospective cohort of healthy infants, maternal GWG was associated with differences in the composition of infant fecal microbiota. Particularly, higher GWG was associated with a lower likelihood of a Bacteroides-dominant microbiota profile, as well as lower bacterial community richness and Shannon diversity index. Although the clinical significance of this association is not yet known, this study prompts further investigation into the mechanism through which GWG modulates the infant fecal microbiota and the metabolic implications of distinct microbiota profiles in infants. Understanding how prenatal environmental and behavioral factors shape the early infant fecal microbiota, and how these microbiota affect child health outcomes, may help us develop strategies (e.g., microbiome modulation) to promote child health during pregnancy.
Supplementary Material
Supplemental Table 1. Patient and Fecal Microbiota Characteristics in the Analytic and non-Analytic Cohort.
Supplemental Table 2. Unadjusted Associations between Gestational Weight Gain During Pregnancy and Fecal Microbiota Profiles in Infants, According to Feeding Status (n=84)
Supplemental Figure 1. Correlation of maternal weight gain during pregnancy with bacterial community richness and Shannon diversity index of infant fecal microbiota.
Spearman’s correlation and Pearson’s correlation were used to examine the association of maternal weight gain during pregnancy with infant fecal bacterial community richness (number of genera) and Shannon diversity index, respectively. A) There was a negative correlation between maternal weight gain during pregnancy and bacterial community richness (Spearman’s rho=−0.25, P=0.02). B) There was a negative correlation between maternal weight gain during pregnancy and Shannon diversity index (Pearson’s rho=−0.25, P=0.02).
What is known.
Fecal microbiota during infancy is shaped by several prenatal and postpartum factors.
Gestational weight gain appears to be associated with child health outcomes.
The association between gestational weight gain and early infant microbiota is not known.
What is new.
Maternal gestational weight gain is associated with infant fecal microbiota profiles, bacterial community richness, and Shannon diversity index.
This study prompts further investigation into the potential mechanisms by which gestational weight gain modulates infant fecal microbiota and how these variations in microbiota correlate with long term health outcomes.
Acknowledgments
Source of Funding: This work was supported by the grants P30DK040561, UG3 OD-023253, U01 AI-087881, R01 AI-114552, R21 HL-129909, and R25DK103579 from the National Institutes of Health (Bethesda, MD). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Dr. Fiechtner’s time was supported by grant number K12HS022986 from the Agency for Healthcare Research and Quality. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality.
Footnotes
Conflicts of Interest: Drs. Ajami and Petrosino own shares at Diversigen Inc., a microbiome research company. The remaining authors report no conflicts of interest.
References
- 1.Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977;34:529–33. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 5.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. The role of Gut Microbiota in the development of obesity and Diabetes. Lipids Health Dis. 2016;15:108. doi: 10.1186/s12944-016-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–50. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 7.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 9.Reinhardt C, Reigstad CS, Backhed F. Intestinal microbiota during infancy and its implications for obesity. J Pediatr Gastroenterol Nutr. 2009;48:249–56. doi: 10.1097/mpg.0b013e318183187c. [DOI] [PubMed] [Google Scholar]
- 10.Harpsoe MC, Basit S, Bager P, et al. Maternal obesity, gestational weight gain, and risk of asthma and atopic disease in offspring: a study within the Danish National Birth Cohort. J Allergy Clin Immunol. 2013;131:1033–40. doi: 10.1016/j.jaci.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Dello Russo M, Ahrens W, De Vriendt T, et al. Gestational weight gain and adiposity, fat distribution, metabolic profile, and blood pressure in offspring: the IDEFICS project. Int J Obes (Lond) 2013;37:914–9. doi: 10.1038/ijo.2013.35. [DOI] [PubMed] [Google Scholar]
- 12.Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother’s weight on infant’s microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. 2010;92:1023–30. doi: 10.3945/ajcn.2010.29877. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa K, Linnemann RW, Avadhanula V, et al. Detection of respiratory syncytial virus and rhinovirus in healthy infants. BMC Res Notes. 2015;8:718. doi: 10.1186/s13104-015-1695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa K, Linneman RW, Mansbach JM, Ajami NJ, Espinola JA, Fiechtner LG, Petrosino JF, Camargo CA., Jr Household siblings and nasal and fecal microbiota in infants. Pediatr Int. 2016 Sep; doi: 10.1111/ped.13168. [e-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansbach JM, Hasegawa K, Henke DM, et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol. 2016;137:1909–13 e4. doi: 10.1016/j.jaci.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camargo CA., Jr Gestational weight gain and offspring asthma: a novel opportunity for primary prevention research. Clin Exp Allergy. 2015;45:544–6. doi: 10.1111/cea.12436. [DOI] [PubMed] [Google Scholar]
- 17.Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557–78. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa K, Linnemann RW, Mansbach JM, et al. The Fecal Microbiota Profile and Bronchiolitis in Infants Pediatrics. 2016;138 doi: 10.1542/peds.2016-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Human Microbiome Project C. A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 22.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufman L, Rousseeuw PJ. Partitioning around medoids (program pam) Finding groups in data: an introduction to cluster analysis. 1990:68–125. [Google Scholar]
- 24.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2001;63:411–23. [Google Scholar]
- 25.Urwin HJ, Miles EA, Noakes PS, et al. Effect of salmon consumption during pregnancy on maternal and infant faecal microbiota, secretory IgA and calprotectin. Br J Nutr. 2014;111:773–84. doi: 10.1017/S0007114513003097. [DOI] [PubMed] [Google Scholar]
- 26.Santacruz A, Collado MC, Garcia-Valdes L, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010;104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 27.Leermakers ET, Sonnenschein-van der Voort AM, Gaillard R, et al. Maternal weight, gestational weight gain and preschool wheezing: the Generation R Study. Eur Respir J. 2013;42:1234–43. doi: 10.1183/09031936.00148212. [DOI] [PubMed] [Google Scholar]
- 28.Schack-Nielsen L, Michaelsen KF, Gamborg M, Mortensen EL, Sorensen TI. Gestational weight gain in relation to offspring body mass index and obesity from infancy through adulthood. Int J Obes (Lond) 2010;34:67–74. doi: 10.1038/ijo.2009.206. [DOI] [PubMed] [Google Scholar]
- 29.Chu DM, Antony KM, Ma J, et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016;8:77. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcobal A, Barboza M, Sonnenburg ED, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–14. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riva A, Borgo F, Lassandro C, et al. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ Microbiol. 2016 doi: 10.1111/1462-2920.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjorksten B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy. 1999;29:342–6. doi: 10.1046/j.1365-2222.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 33.Vael C, Verhulst SL, Nelen V, Goossens H, Desager KN. Intestinal microflora and body mass index during the first three years of life: an observational study. Gut Pathog. 2011;3:8. doi: 10.1186/1757-4749-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeBouder E, Rey-Nores JE, Raby AC, et al. Modulation of neonatal microbial recognition: TLR-mediated innate immune responses are specifically and differentially modulated by human milk. J Immunol. 2006;176:3742–52. doi: 10.4049/jimmunol.176.6.3742. [DOI] [PubMed] [Google Scholar]
- 35.Kostic AD, Gevers D, Siljander H, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–73. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–81. e1–3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin VJ, Leonard MM, Fiechtner L, Fasano A. Transitioning From Descriptive to Mechanistic Understanding of the Microbiome: The Need for a Prospective Longitudinal Approach to Predicting Disease. J Pediatr. 2016 doi: 10.1016/j.jpeds.2016.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romano-Keeler J, Weitkamp JH. Maternal influences on fetal microbial colonization and immune development. Pediatr Res. 2015;77:189–95. doi: 10.1038/pr.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nutrition During Pregnancy: Part I Weight Gain: Part II Nutrient Supplements. Washington (DC): 1990. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Patient and Fecal Microbiota Characteristics in the Analytic and non-Analytic Cohort.
Supplemental Table 2. Unadjusted Associations between Gestational Weight Gain During Pregnancy and Fecal Microbiota Profiles in Infants, According to Feeding Status (n=84)
Supplemental Figure 1. Correlation of maternal weight gain during pregnancy with bacterial community richness and Shannon diversity index of infant fecal microbiota.
Spearman’s correlation and Pearson’s correlation were used to examine the association of maternal weight gain during pregnancy with infant fecal bacterial community richness (number of genera) and Shannon diversity index, respectively. A) There was a negative correlation between maternal weight gain during pregnancy and bacterial community richness (Spearman’s rho=−0.25, P=0.02). B) There was a negative correlation between maternal weight gain during pregnancy and Shannon diversity index (Pearson’s rho=−0.25, P=0.02).


