Figure 1. Analysis of main chain conformations of phosphopeptides.
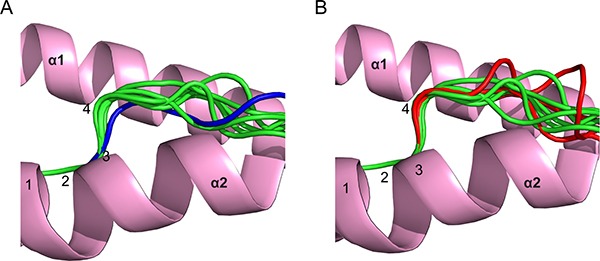
(A) Comparison of peptide main-chain conformation around position 4 for a canonical phosphopeptide derived from insulin receptor substrate 2 (coloured blue; PDB code 3FQX obtained from a previous study by Petersen et al. [12]), relative to other canonical phosphopeptide structures from previous studies [12, 15] (green). (B) Comparison of main chain conformations of PKD2p, RQA_Vp, and RQIp (all shown in red), relative to previously solved canonical phosphopeptides (shown in green). All superpositions were based on Cα atoms of residues at positions 1–3.
