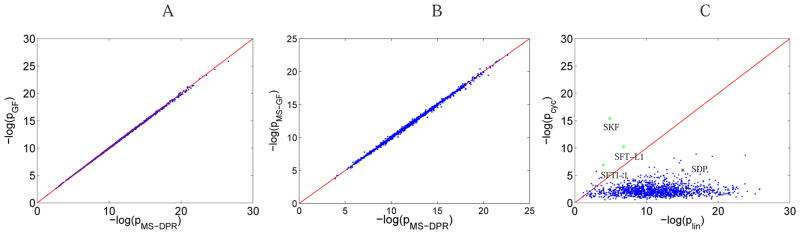
Figure 7.
(A) Comparison of −log10 of generating function p-value with MS-DPR p-value for 1388 peptides from ISB database. Red line shows the x = y line. Correlation between the two p-values is 0.9998. Non-standard amino acid model is used, assuming each peptide has a fixed known length, and peak count score. MS-GF approach17 is modified accordingly, to satisfy these assumptions. (B) Comparison of −log10 of the original, publicly available MS-GF p-value with MS-DPR p-value. Correlation between the two p-values is 0.9990. Standard amino acid model is used, with the variable peptide length assumption and MS-GF score. 17 (C) Comparison of −log10 of plin, versus −log10 of pcyc for SFTI-1, SFT-L2, SKF, SDP, and spectra from the ISB dataset. Cyclic peptides SFTI-1, SFT-L2 and SKF are shown as green stars, and linear peptide SDP is shown as a black star. Blue dots show spectra from ISB dataset, and red line shows the x = y line.