Figure 13.
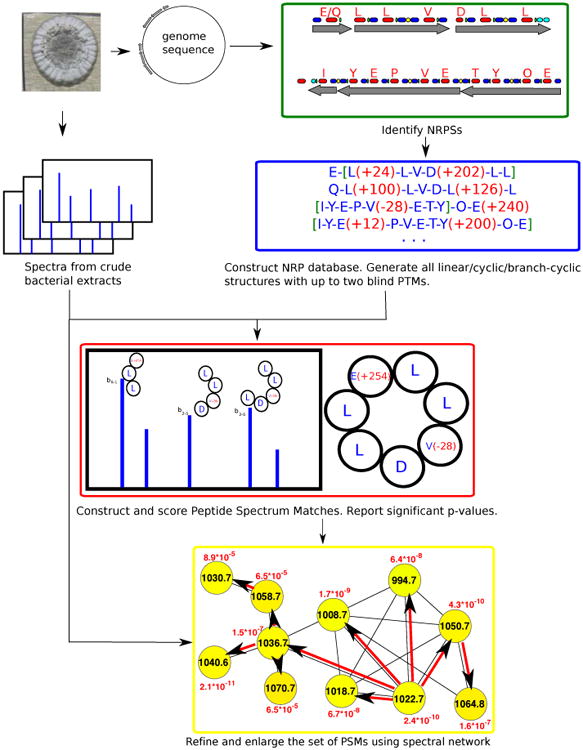
NRPquest pipeline starts with with mining the microbial genome for putative NRPs using NRPSpredictor2 [40] and Antismash [72]. Two blind modifications are added to each NRP and all possible structures (linear/cyclic/branch-cyclic) are considered. PSMs are formed between each spectrum and each putative modified NRP with feasible mass difference. PSMs are scored, and significant PSMs are further analysed by spectral networks. PSMs are rescored based on how well other nodes in their spectral network cluster (connected component) are explained [27].
