Figure 5.
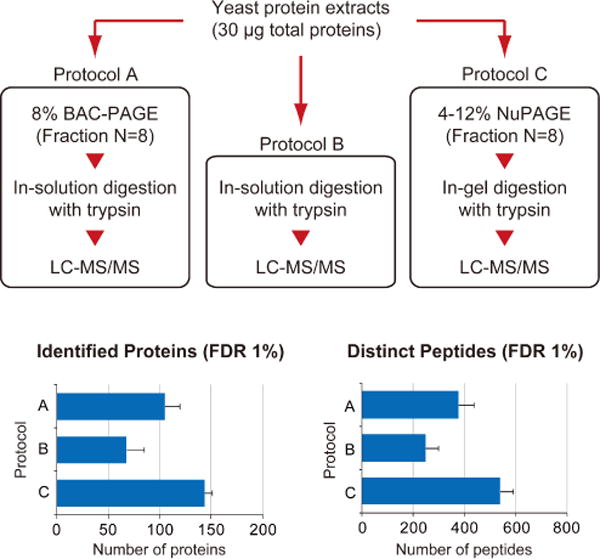
Comparative evaluation of different sample treatments for bottom-up proteomics. After tryptic digestion of a yeast protein extract (Promega) using different three methods independently, the digested peptides were subjected to LC-MS/MS analysis. For protein identification, MS/MS spectra were searched against the UniProt yeast proteome database by ProteinPilot using the following parameters: cys alkylation, iodoacetamide; digestion, trypsin; processing parameters, biological modification; and search effort, through ID. The values represent the averages from three independent experiments ± S.D.
