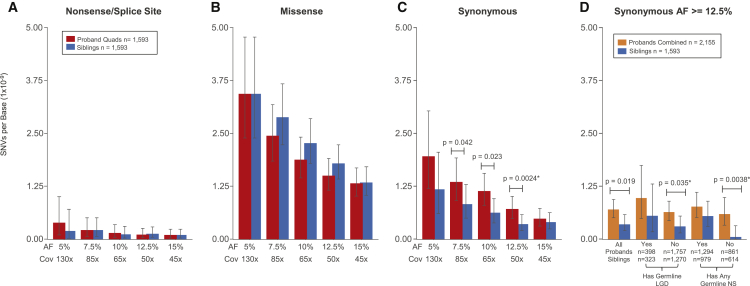
Figure 2.
Rates and Burden of SNV PMMs in the Simons Simplex Collection (SSC)
(A–C) Rates and burden analyses of PMMs in quad families of the SSC. Mean rates with 95% Poisson CIs (exact method) are shown.
(A) Nonsense/splice PMM rates are similar and not evaluated further given their low frequency.
(B) Missense PMMs show no evidence of burden in probands from quad families.
(C) Synonymous PMMs show an unexpected burden in probands from quad families. Significance determined using a two-sided Wilcoxon signed-rank test. ∗FDR < 0.05 using the Benjamini-Yekutieli approach.
(D) Analysis of synonymous PMMs at AF 12.5%-50× in the full SSC and subcohorts. Mean rates with 95% Poisson CIs (exact method) are shown for combined probands (quad + trio families) and unaffected siblings. Abbreviations are as follows: SSC subcohorts all, all families within the cohort passing quality criteria; Has Germline LGD, denotes whether or not proband in family has a LGD GDM or gene disrupting de novo CNV; Has Any Germline NS, denotes whether or not proband in family has any NS GDM (includes the LGD set). Significance determined using a two-sided Wilcoxon rank sum test. ∗FDR < 0.05 using the Benjamini-Yekutieli approach.