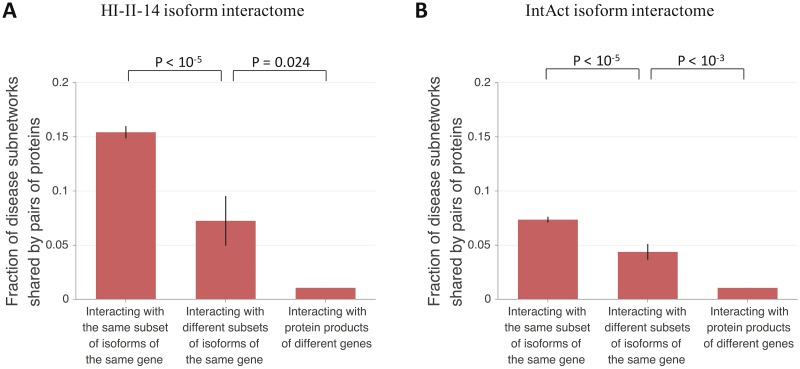
Fig 3. Fraction of shared disease subnetworks for protein pairs with different types of isoform interaction profiles.
(A) Fraction of shared disease subnetworks for pairs of proteins interacting with the same subset of isoforms of the same gene, with different subsets of isoforms of the same gene, and with protein products of different genes in the predicted HI-II-14 isoform interactome. (B) Fraction of shared disease subnetworks for pairs of proteins interacting with the same subset of isoforms of the same gene, with different subsets of isoforms of the same gene, and with protein products of different genes in the predicted IntAct isoform interactome. Fraction of shared disease subnetworks was calculated using the Jaccard similarity index. Only pairs in which both proteins are associated with at least one disease subnetwork were considered. A protein belongs to a disease subnetwork if that protein or any of its first-degree neighbors in the HI-II-14 reference binary interactome are associated with the disease. Error bars represent standard errors of the mean. Statistical significance was calculated using a two-sided bootstrap test.