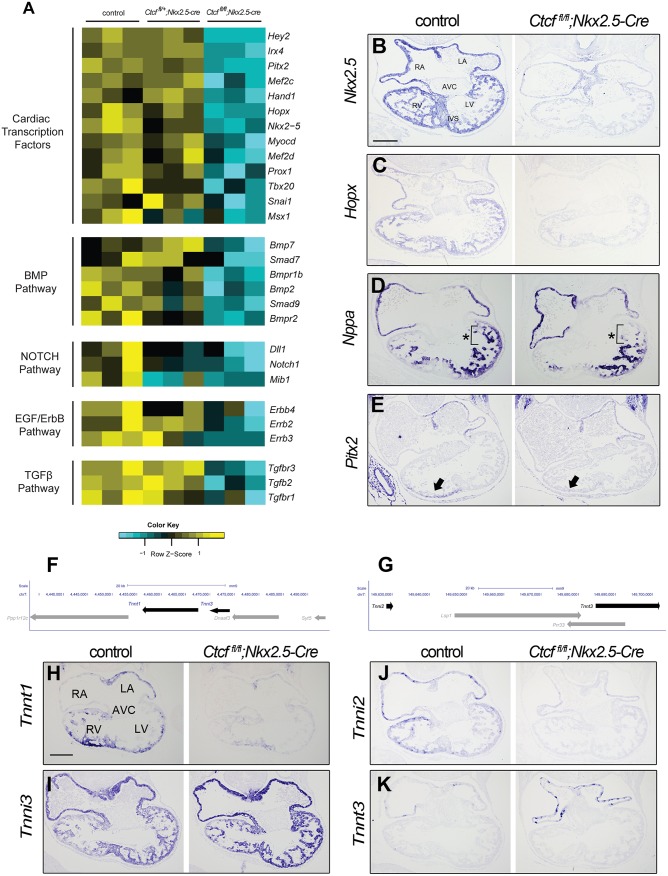
Fig 4. The cardiac developmental program is abrogated in Ctcf mutant hearts.
(A) Deletion of Ctcf leads to downregulation of genes encoding key cardiac developmental transcription factors and signaling-pathway components. The heatmap shows normalized gene expression values from the RNA-seq data for controls (Ctcffl/+), heterozygotes (Ctcffl/+;Nkx2.5-Cre) and homozygotes (Ctcffl/fl;Nkx2.5-Cre) across three biological replicates each. (B-E) In situ hybridization in sections of E10.5 control and mutant (Ctcffl/fl;Nkx2-5-Cre) hearts for Nkx2.5 (B), Hopx (C), which are strongly downregulated; and Nppa (D) and Pitx2 (E), which lose expression in the left ventricle (brackets and asterisks in D) or right ventricle (arrow in E), respectively. (F, G) Genomic region containing Tnnt1 and Tnni3 (F; mm9, chr7:4,433,080–4,493,651) or Tnni2 and Tnnt3 (G; mm9, chr7:149,623,608–149,703,181). Troponin genes are highlighted in black. (H-K) In situ hybridization in sections of E10.5 control and mutant hearts for Tnnt1 (H), Tnni3 (I), Tnni2 (J) and Tnnt3 (K). RA, right atria; LA, left atria; RV, right ventricle; LV, left ventricle; AVC, atrioventricular canal; IVS, interventricular septum. Scale bar, 200 μm.